Molecular breakpoint cloning and gene expression studies of a novel translocation t(4;15)(q27;q11.2) associated with Prader-Willi syndrome
- PMID: 15877813
- PMCID: PMC1142316
- DOI: 10.1186/1471-2350-6-18
Molecular breakpoint cloning and gene expression studies of a novel translocation t(4;15)(q27;q11.2) associated with Prader-Willi syndrome
Abstract
Background: Prader-Willi syndrome (MIM #176270; PWS) is caused by lack of the paternally-derived copies, or their expression, of multiple genes in a 4 Mb region on chromosome 15q11.2. Known mechanisms include large deletions, maternal uniparental disomy or mutations involving the imprinting center. De novo balanced reciprocal translocations in 5 reported individuals had breakpoints clustering in SNRPN intron 2 or exon 20/intron 20. To further dissect the PWS phenotype and define the minimal critical region for PWS features, we have studied a 22 year old male with a milder PWS phenotype and a de novo translocation t(4;15)(q27;q11.2).
Methods: We used metaphase FISH to narrow the breakpoint region and molecular analyses to map the breakpoints on both chromosomes at the nucleotide level. The expression of genes on chromosome 15 on both sides of the breakpoint was determined by RT-PCR analyses.
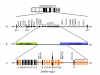
Results: Pertinent clinical features include neonatal hypotonia with feeding difficulties, hypogonadism, short stature, late-onset obesity, learning difficulties, abnormal social behavior and marked tolerance to pain, as well as sticky saliva and narcolepsy. Relative macrocephaly and facial features are not typical for PWS. The translocation breakpoints were identified within SNRPN intron 17 and intron 10 of a spliced non-coding transcript in band 4q27. LINE and SINE sequences at the exchange points may have contributed to the translocation event. By RT-PCR of lymphoblasts and fibroblasts, we find that upstream SNURF/SNRPN exons and snoRNAs HBII-437 and HBII-13 are expressed, but the downstream snoRNAs PWCR1/HBII-85 and HBII-438A/B snoRNAs are not.
Conclusion: As part of the PWCR1/HBII-85 snoRNA cluster is highly conserved between human and mice, while no copy of HBII-438 has been found in mouse, we conclude that PWCR1/HBII-85 snoRNAs is likely to play a major role in the PWS- phenotype.
Figures

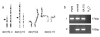




References
-
- Cassidy SB. Prader-Willi syndrome. 2001. pp. 301–322.
-
- Ledbetter DH, Riccardi VM, Airhart SD, Strobel RJ, Keenan BS, Crawford JD. Deletions of chromosome 15 as a cause of the Prader-Willi syndrome. N Engl J Med. 1981;304:325–329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

