A combined approach exploring gene function based on worm-human orthology
- PMID: 15877817
- PMCID: PMC1112593
- DOI: 10.1186/1471-2164-6-65
A combined approach exploring gene function based on worm-human orthology
Abstract
Background: Many aspects of the nematode Caenorhabditis elegans biology are conserved between invertebrates and vertebrates establishing this particular organism as an excellent genetic model. Because of its small size, large populations and self-fertilization of the hermaphrodite, functional predictions carried out by genetic modifications as well as RNAi screens, can be rapidly tested.
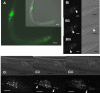
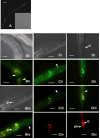
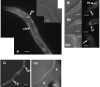
Results: In order to explore the function of a set of C. elegans genes of unknown function, as well as their potential functional roles in the human genome, we performed a phylogenetic analysis to select the most probable worm orthologs. A total of 13 C. elegans genes were subjected to down-regulation via RNAi and characterization of expression profiles using GFP strains. Previously unknown distinct expression patterns were observed for four of the analyzed genes, as well as four visible RNAi phenotypes. In addition, subcellular protein over-expression profiles of the human orthologs for seven out of the thirteen genes using human cells were also analyzed.
Conclusion: By combining a whole-organism approach using C. elegans with complementary experimental work done on human cell lines, this analysis extends currently available information on the selected set of genes.
Figures



References
-
- McKay SJ, Johnsen R, Khattra J, Asano J, Baillie DL, Chan S, Dube N, Fang L, Goszczynski B, Ha E, Halfnight E, Hollebakken R, Huang P, Hung K, Jensen V, Jones SJ, Kai H, Li D, Mah A, Marra M, McGhee J, Newbury R, Pouzyrev A, Riddle DL, Sonnhammer E, Tian H, Tu D, Tyson JR, Vatcher G, Warner A, Wong K, Zhao Z, Moerman DG. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb Symp Quant Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. - DOI - PubMed
-
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, Hannak E, Kirkham M, Pichler S, Flohrs K, Goessen A, Leidel S, Alleaume AM, Martin C, Ozlu N, Bork P, Hyman AA. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

