Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry
- PMID: 15878991
- PMCID: PMC1129118
- DOI: 10.1073/pnas.0502164102
Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry
Abstract
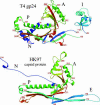
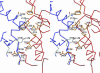
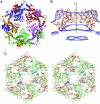
Gene product (gp) 24 of bacteriophage T4 forms the pentameric vertices of the capsid. Using x-ray crystallography, we found the principal domain of gp24 to have a polypeptide fold similar to that of the HK97 phage capsid protein plus an additional insertion domain. Fitting gp24 monomers into a cryo-EM density map of the mature T4 capsid suggests that the insertion domain interacts with a neighboring subunit, effecting a stabilization analogous to the covalent crosslinking in the HK97 capsid. Sequence alignment and genetic data show that the folds of gp24 and the hexamer-forming capsid protein, gp23*, are similar. Accordingly, models of gp24* pentamers, gp23* hexamers, and the whole capsid were built, based on a cryo-EM image reconstruction of the capsid. Mutations in gene 23 that affect capsid shape map to the capsomer's periphery, whereas mutations that allow gp23 to substitute for gp24 at the vertices modify the interactions between monomers within capsomers. Structural data show that capsid proteins of most tailed phages, and some eukaryotic viruses, may have evolved from a common ancestor.
Figures


References
-
- Conway, J. F., Wikoff, W. R., Cheng, N., Duda, R. L., Hendrix, R. W., Johnson, J. E. & Steven, A. C. (2001) Science 292, 744-748. - PubMed
-
- Black, L. W., Showe, M. K. & Steven, A. C. (1994) in Molecular Biology of Bacteriophage T4, ed. Karam, J. D. (Am. Soc. Microbiol., Washington, DC), pp. 218-258.
-
- Driedonks, R. A., Engel, A., tenHeggeler, B. & van Driel, R. (1981) J. Mol. Biol. 152, 641-662. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

