Identification in the ancient protist Giardia lamblia of two eukaryotic translation initiation factor 4E homologues with distinctive functions
- PMID: 15879529
- PMCID: PMC1140097
- DOI: 10.1128/EC.4.5.948-959.2005
Identification in the ancient protist Giardia lamblia of two eukaryotic translation initiation factor 4E homologues with distinctive functions
Abstract
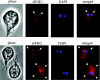
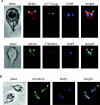
Eukaryotic translation initiation factor 4E (eIF4E) binds to the m(7)GTP of capped mRNAs and is an essential component of the translational machinery that recruits the 40S small ribosomal subunit. We describe here the identification and characterization of two eIF4E homologues in an ancient protist, Giardia lamblia. Using m(7)GTP-Sepharose affinity column chromatography, a specific binding protein was isolated and identified as Giardia eIF4E2. The other homologue, Giardia eIF4E1, bound only to the m(2,2,7)GpppN structure. Although neither homologue can rescue the function of yeast eIF4E, a knockdown of eIF4E2 mRNA in Giardia by a virus-based antisense ribozyme decreased translation, which was shown to use m(7)GpppN-capped mRNA as a template. Thus, eIF4E2 is likely the cap-binding protein in a translation initiation complex. The same knockdown approach indicated that eIF4E1 is not required for translation in Giardia. Immunofluorescence assays showed wide distribution of both homologues in the cytoplasm. But eIF4E1 was also found concentrated and colocalized with the m(2,2,7)GpppN cap, 16S-like rRNA, and fibrillarin in the nucleolus-like structure in the nucleus. eIF4E1 depletion from Giardia did not affect mRNA splicing, but the protein was bound to Giardia small nuclear RNAs D and H known to have an m(2,2,7)GpppN cap, thus suggesting a novel function not yet observed among other eIF4Es in eukaryotes.
Figures






Similar articles
-
Translation initiation factors GleIF4E2 and GleIF4A can interact directly with the components of the pre-initiation complex to facilitate translation initiation in Giardia lamblia.Mol Biochem Parasitol. 2020 Mar;236:111258. doi: 10.1016/j.molbiopara.2020.111258. Epub 2020 Jan 20. Mol Biochem Parasitol. 2020. PMID: 31968220
-
Distinct recruitment of human eIF4E isoforms to processing bodies and stress granules.BMC Mol Biol. 2016 Aug 30;17(1):21. doi: 10.1186/s12867-016-0072-x. BMC Mol Biol. 2016. PMID: 27578149 Free PMC article.
-
Investigating the consequences of eIF4E2 (4EHP) interaction with 4E-transporter on its cellular distribution in HeLa cells.PLoS One. 2013 Aug 21;8(8):e72761. doi: 10.1371/journal.pone.0072761. eCollection 2013. PLoS One. 2013. PMID: 23991149 Free PMC article.
-
eIF4E and Interactors from Unicellular Eukaryotes.Int J Mol Sci. 2020 Mar 21;21(6):2170. doi: 10.3390/ijms21062170. Int J Mol Sci. 2020. PMID: 32245232 Free PMC article. Review.
-
Taking a re-look at cap-binding signatures of the mRNA cap-binding protein eIF4E orthologues in trypanosomatids.Mol Cell Biochem. 2021 Feb;476(2):1037-1049. doi: 10.1007/s11010-020-03970-w. Epub 2020 Nov 10. Mol Cell Biochem. 2021. PMID: 33169189 Review.
Cited by
-
Analysis of Giardia lamblia Nucleolus as Drug Target: A Review.Pharmaceuticals (Basel). 2023 Aug 16;16(8):1168. doi: 10.3390/ph16081168. Pharmaceuticals (Basel). 2023. PMID: 37631082 Free PMC article. Review.
-
The evolution of the Puf superfamily of proteins across the tree of eukaryotes.BMC Biol. 2020 Jun 30;18(1):77. doi: 10.1186/s12915-020-00814-3. BMC Biol. 2020. PMID: 32605621 Free PMC article.
-
Multispecies reconstructions uncover widespread conservation, and lineage-specific elaborations in eukaryotic mRNA metabolism.PLoS One. 2018 Mar 21;13(3):e0192633. doi: 10.1371/journal.pone.0192633. eCollection 2018. PLoS One. 2018. PMID: 29561870 Free PMC article.
-
A reduced VWA domain-containing proteasomal ubiquitin receptor of Giardia lamblia localizes to the flagellar pore regions in microtubule-dependent manner.Parasit Vectors. 2015 Feb 24;8:120. doi: 10.1186/s13071-015-0737-1. Parasit Vectors. 2015. PMID: 25888841 Free PMC article.
-
snoRNA, a novel precursor of microRNA in Giardia lamblia.PLoS Pathog. 2008 Nov;4(11):e1000224. doi: 10.1371/journal.ppat.1000224. Epub 2008 Nov 28. PLoS Pathog. 2008. PMID: 19043559 Free PMC article.
References
-
- Adam, R. D. 2000. The Giardia lamblia genome. Int. J. Parasitol. 30:475-484. - PubMed
-
- Altmann, M., P. P. Muller, J. Pelletier, N. Sonenberg, and H. Trachsel. 1989. A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J. Biol. Chem. 264:12145-12147. - PubMed
-
- Carberry, S. E., R. E. Rhoads, and D. J. Goss. 1989. A spectroscopic study of the binding of m7GTP and m7GpppG to human protein synthesis initiation factor 4E. Biochemistry 28:8078-8083. - PubMed
-
- Clauser, K. R., P. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (±10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871-2882. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

