p25alpha is flexible but natively folded and binds tubulin with oligomeric stoichiometry
- PMID: 15883183
- PMCID: PMC2253386
- DOI: 10.1110/ps.041285605
p25alpha is flexible but natively folded and binds tubulin with oligomeric stoichiometry
Abstract
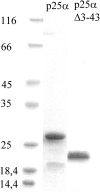
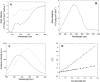
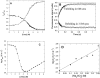
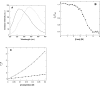
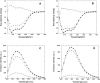
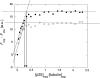
p25alpha is a 219-residue protein which stimulates aberrant tubulin polymerization and is implicated in a variety of other functions. The protein has unusual secondary structure involving significant amounts of random coil, and binding to microtubules is accompanied by a large structural change, suggesting a high degree of plasticity. p25alpha has been proposed to be natively unfolded, so that folding is coupled to interaction with its physiological partners. Here we show that recombinant human p25alpha is folded under physiological conditions, since it has a well structured and solvent-sequestered aromatic environment and considerable chemical shift dispersion of amide and aliphatic protons. With increasing urea concentrations, p25alpha undergoes clear spectral changes suggesting significant loss of structure. p25alpha unfolds cooperatively in urea according to a simple two-state transition with a stability in water of approximately 5 kcal/mol. The protein behaves as a monomer and refolds with a transient on-pathway folding intermediate. However, high sensitivity to proteolytic attack and abnormal gel filtration migration behavior suggests a relatively extended structure, possibly organized in distinct domains. A deletion mutant of p25alpha lacking residues 3-43 also unfolds cooperatively and with similar stability, suggesting that the N-terminal region is largely unstructured. Both proteins undergo significant loss of structure when bound to monomeric tubulin. The stoichiometry of binding is estimated to be 3-4 molecules of tubulin per p25alpha and is not significantly affected by the deletion of residues 3-43. In conclusion, we dismiss the proposal that p25alpha is natively unfolded, although the protein is relatively flexible. This flexibility may be linked to its tubulin-binding properties.
Figures
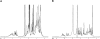
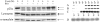
Similar articles
-
The expression of tubulin polymerization promoting protein TPPP/p25alpha is developmentally regulated in cultured rat brain oligodendrocytes and affected by proteolytic stress.Glia. 2008 Dec;56(16):1736-46. doi: 10.1002/glia.20720. Glia. 2008. PMID: 18563798
-
P25alpha/Tubulin polymerization promoting protein expression by myelinating oligodendrocytes of the developing rat brain.J Neurochem. 2006 Oct;99(1):333-42. doi: 10.1111/j.1471-4159.2006.04073.x. Epub 2006 Jul 31. J Neurochem. 2006. PMID: 16879710
-
Multiple roles of heparin in the aggregation of p25α.J Mol Biol. 2012 Aug 24;421(4-5):601-15. doi: 10.1016/j.jmb.2012.01.050. Epub 2012 Feb 3. J Mol Biol. 2012. PMID: 22326478
-
Tubulin rings: which way do they curve?Curr Opin Struct Biol. 2003 Apr;13(2):256-61. doi: 10.1016/s0959-440x(03)00029-0. Curr Opin Struct Biol. 2003. PMID: 12727521 Review.
-
Tubulin structure: insights into microtubule properties and functions.Curr Opin Struct Biol. 1998 Dec;8(6):785-91. doi: 10.1016/s0959-440x(98)80099-7. Curr Opin Struct Biol. 1998. PMID: 9914260 Review.
Cited by
-
An unusual intrinsically disordered protein from the model legume Lotus japonicus stabilizes proteins in vitro.J Biol Chem. 2008 Nov 7;283(45):31142-52. doi: 10.1074/jbc.M805024200. Epub 2008 Sep 8. J Biol Chem. 2008. PMID: 18779323 Free PMC article.
-
Construct optimization for protein NMR structure analysis using amide hydrogen/deuterium exchange mass spectrometry.Proteins. 2009 Sep;76(4):882-94. doi: 10.1002/prot.22394. Proteins. 2009. PMID: 19306341 Free PMC article.
-
Neurons and Glia Interplay in α-Synucleinopathies.Int J Mol Sci. 2021 May 8;22(9):4994. doi: 10.3390/ijms22094994. Int J Mol Sci. 2021. PMID: 34066733 Free PMC article. Review.
-
p25alpha relocalizes in oligodendroglia from myelin to cytoplasmic inclusions in multiple system atrophy.Am J Pathol. 2007 Oct;171(4):1291-303. doi: 10.2353/ajpath.2007.070201. Epub 2007 Sep 6. Am J Pathol. 2007. PMID: 17823288 Free PMC article.
-
Identification of synaptosomal proteins binding to monomeric and oligomeric α-synuclein.PLoS One. 2015 Feb 6;10(2):e0116473. doi: 10.1371/journal.pone.0116473. eCollection 2015. PLoS One. 2015. PMID: 25659148 Free PMC article.
References
-
- Andrade, M.A., Chacón, P., Merelo, J.J., and Morán, F. 1993. Evaluation of secondary structure of proteins from UV circular dichroism using an unsupervised learning neural network. Protein Eng. 6 383–390. - PubMed
-
- Baldwin, R. 1996. On-pathway versus off-pathway folding intermediates. Fold. Des. 1 R1–R8. - PubMed
-
- Buck, M., Radford, S.E., and Dobson, C.M. 1993. A partially folded state of hen egg white lysozyme in trifluoroethanol: Structural characterization and implications for protein folding. Biochemistry 32 669–678. - PubMed
-
- Capaldi, A.P., Shastry, M.C., Kleanthous, C., Roder, H., and Radford, S.E. 2001. Ultrarapid mixing experiments reveal that Im7 folds via an on-pathway intermediate. Nat. Struct. Biol. 8 68–72. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

