Novel and economical purification of recombinant proteins: intein-mediated protein purification using in vivo polyhydroxybutyrate (PHB) matrix association
- PMID: 15883185
- PMCID: PMC2253394
- DOI: 10.1110/ps.041296305
Novel and economical purification of recombinant proteins: intein-mediated protein purification using in vivo polyhydroxybutyrate (PHB) matrix association
Abstract
This work combines two well-established technologies to generate a breakthrough in protein production and purification. The first is the production of polyhydroxybutyrate (PHB) granules in engineered strains of Escherichia coli. The second is a recently developed group of self-cleaving affinity tags based on protein splicing elements known as inteins. By combining these technologies with a PHB-specific binding protein, a self-contained protein expression and purification system has been developed. In this system, the PHB-binding protein effectively acts as an affinity tag for desired product proteins. The tagged product proteins are expressed in E. coli strains that also produce intracellular PHB granules, where they bind to the granules via the PHB-binding tag. The granules and attached proteins can then be easily recovered following cell lysis by simple mechanical means. Once purified, the product protein is self-cleaved from the granules and released into solution in a substantially purified form. This system has been successfully used at laboratory scale to purify several active test proteins at reasonable yield. By allowing the bacterial cells to effectively produce both the affinity resin and tagged target protein, the cost associated with the purification of recombinant proteins could be greatly reduced. It is expected that this combination of improved economics and simplicity will constitute a significant breakthrough in both large-scale production of purified proteins and enzymes and high-throughput proteomics studies of peptide libraries.
Figures


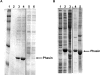

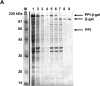


Comment in
-
Proteins from PHB granules.Protein Sci. 2005 Jun;14(6):1385-6. doi: 10.1110/ps.051418305. Protein Sci. 2005. PMID: 15929993 Free PMC article. No abstract available.
References
-
- Ausubel, F.M. 1998. Protein analysis. In Current protocols in molecular biology (eds. F.M. Ausubel et al.), Section 10.11.14. John Wiley & Sons, New York.
-
- Chong, S., Mersha, F.B., Comb, D.G., Scott, M.E., Landry, D., Vence, L.M., Perler, F.B., Benner, J., Kucera, R.B., Hirvonen, C.A., et al. 1997. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192 271–281. - PubMed
-
- Doi, Y. 1990. Fermentation and analysis of microbial polyesters. In Microbial polyesters, p. 156. VCH, New York.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

