Insights into TOR function and rapamycin response: chemical genomic profiling by using a high-density cell array method
- PMID: 15883373
- PMCID: PMC1091748
- DOI: 10.1073/pnas.0500297102
Insights into TOR function and rapamycin response: chemical genomic profiling by using a high-density cell array method
Erratum in
- Proc Natl Acad Sci U S A. 2006 Jun 13;103(24):9374
- Proc Natl Acad Sci U S A. 2006 Sep 5;103(36):13560
Abstract
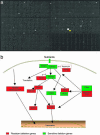
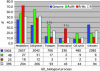
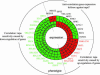
With the advent of complete genome sequences, large-scale functional analyses are generating new excitement in biology and medicine. To facilitate genomewide functional analyses, we developed a high-density cell array with quantitative and automated readout of cell fitness. Able to print at > x 10 higher density on a standard microtiter plate area than currently possible, our cell array allows single-plate screening of the complete set of Saccharomyces cerevisiae gene-deletion library and significantly reduces the amount of small molecules and other materials needed for the study. We used this method to map the relation between genes and cell fitness in response to rapamycin, a medically important natural product that targets the eukaryotic kinase Tor. We discuss the implications for pharmacogenomics and the uncharted complexity in genotype-dependent drug response in molecularly targeted therapies. Our analysis leads to several basic findings, including a class of gene deletions that confer better fitness in the presence of rapamycin. This result provides insights into possible therapeutic uses of rapamycin/CCI-779 in the treatment of neurodegenerative diseases (including Alzheimer's, Parkinson's, and Huntington's diseases), and cautions the possible existence of similar rapamycin-enhanceable mutations in cancer. It is well established in yeast that although TOR2 has a unique rapamycin-insensitive function, TOR1 and TOR2 are interchangeable in the rapamycin-sensitive functions. We show that even the rapamycin-sensitive functions are distinct between TOR1 and TOR2 and map the functional difference to a approximately 120-aa region at the N termini of the proteins. Finally, we discuss using cell-based genomic pattern recognition in designing electronic or optical biosensors.
Figures

Similar articles
-
Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae.Genetics. 2007 Aug;176(4):2139-50. doi: 10.1534/genetics.107.072835. Epub 2007 Jun 11. Genetics. 2007. PMID: 17565946 Free PMC article.
-
Rapamycin inhibits yeast nucleotide excision repair independently of tor kinases.Toxicol Sci. 2010 Jan;113(1):77-84. doi: 10.1093/toxsci/kfp238. Epub 2009 Oct 5. Toxicol Sci. 2010. PMID: 19805410 Free PMC article.
-
Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control.Mol Cell. 2002 Sep;10(3):457-68. doi: 10.1016/s1097-2765(02)00636-6. Mol Cell. 2002. PMID: 12408816
-
Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae.Microbiol Mol Biol Rev. 2002 Dec;66(4):579-91, table of contents. doi: 10.1128/MMBR.66.4.579-591.2002. Microbiol Mol Biol Rev. 2002. PMID: 12456783 Free PMC article. Review.
-
Control of translation by the target of rapamycin proteins.Prog Mol Subcell Biol. 2001;27:143-74. doi: 10.1007/978-3-662-09889-9_6. Prog Mol Subcell Biol. 2001. PMID: 11575159 Review. No abstract available.
Cited by
-
FitSearch: a robust way to interpret a yeast fitness profile in terms of drug's mode-of-action.BMC Genomics. 2013;14 Suppl 1(Suppl 1):S6. doi: 10.1186/1471-2164-14-S1-S6. Epub 2013 Jan 21. BMC Genomics. 2013. PMID: 23368702 Free PMC article.
-
Fermentation of high concentrations of maltose by Saccharomyces cerevisiae is limited by the COMPASS methylation complex.Appl Environ Microbiol. 2006 Nov;72(11):7176-82. doi: 10.1128/AEM.01704-06. Epub 2006 Sep 15. Appl Environ Microbiol. 2006. PMID: 16980427 Free PMC article.
-
Yeast as a tool to identify anti-aging compounds.FEMS Yeast Res. 2018 Sep 1;18(6):foy020. doi: 10.1093/femsyr/foy020. FEMS Yeast Res. 2018. PMID: 29905792 Free PMC article. Review.
-
Rapamycin response in tumorigenic and non-tumorigenic hepatic cell lines.PLoS One. 2009 Oct 9;4(10):e7373. doi: 10.1371/journal.pone.0007373. PLoS One. 2009. PMID: 19816606 Free PMC article.
-
Pseudohyphal differentiation defect due to mutations in GPCR and ammonium signaling is suppressed by low glucose concentration: a possible integrated role for carbon and nitrogen limitation.Curr Genet. 2008 Aug;54(2):71-81. doi: 10.1007/s00294-008-0202-1. Epub 2008 Jul 12. Curr Genet. 2008. PMID: 18622617
References
-
- Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. - PubMed
-
- Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387-391. - PubMed
-
- Que, Q. Q. & Winzeler, E. A. (2002) Funct. Integr. Genomics 2, 193-198. - PubMed
-
- Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. (2000) Nature 403, 623-627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

