Integration of the Gene Ontology into an object-oriented architecture
- PMID: 15885145
- PMCID: PMC1156866
- DOI: 10.1186/1471-2105-6-113
Integration of the Gene Ontology into an object-oriented architecture
Abstract
Background: To standardize gene product descriptions, a formal vocabulary defined as the Gene Ontology (GO) has been developed. GO terms have been categorized into biological processes, molecular functions, and cellular components. However, there is no single representation that integrates all the terms into one cohesive model. Furthermore, GO definitions have little information explaining the underlying architecture that forms these terms, such as the dynamic and static events occurring in a process. In contrast, object-oriented models have been developed to show dynamic and static events. A portion of the TGF-beta signaling pathway, which is involved in numerous cellular events including cancer, differentiation and development, was used to demonstrate the feasibility of integrating the Gene Ontology into an object-oriented model.
Results: Using object-oriented models we have captured the static and dynamic events that occur during a representative GO process, "transforming growth factor-beta (TGF-beta) receptor complex assembly" (GO:0007181).
Conclusion: We demonstrate that the utility of GO terms can be enhanced by object-oriented technology, and that the GO terms can be integrated into an object-oriented model by serving as a basis for the generation of object functions and attributes.
Figures

 ) at the end indicate composition. These are read from the diamond end, for example, as 'cell' (GO:0005623) contains a 'membrane' (GO:0016020). Lines with open triangles (
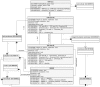
) at the end indicate composition. These are read from the diamond end, for example, as 'cell' (GO:0005623) contains a 'membrane' (GO:0016020). Lines with open triangles ( ) represent generalizations. These are read from the triangle end, as a 'membrane' (GO:0016020) is a general type of 'plasma membrane' (GO:0005886). A) Directed acyclic graph for the cellular component GO terms associated with TGF-beta receptor complex assembly (GO:0007181). B) Object-oriented representation of the DAG described in Figure 1A (white). Additional cellular components not represented by the current Gene Ontology, but essential to the TGF-beta receptor complex assembly process are shown in the gray boxes (i.e. TGF-beta, TGF-beta receptors, SMAD2)
) represent generalizations. These are read from the triangle end, as a 'membrane' (GO:0016020) is a general type of 'plasma membrane' (GO:0005886). A) Directed acyclic graph for the cellular component GO terms associated with TGF-beta receptor complex assembly (GO:0007181). B) Object-oriented representation of the DAG described in Figure 1A (white). Additional cellular components not represented by the current Gene Ontology, but essential to the TGF-beta receptor complex assembly process are shown in the gray boxes (i.e. TGF-beta, TGF-beta receptors, SMAD2)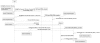

Similar articles
-
An object-oriented programming system for the integration of internet-based bioinformatics resources.Appl Bioinformatics. 2006;5(1):29-39. doi: 10.2165/00822942-200605010-00004. Appl Bioinformatics. 2006. PMID: 16539535
-
GeneKeyDB: a lightweight, gene-centric, relational database to support data mining environments.BMC Bioinformatics. 2005 Mar 24;6:72. doi: 10.1186/1471-2105-6-72. BMC Bioinformatics. 2005. PMID: 15790402 Free PMC article.
-
MILANO--custom annotation of microarray results using automatic literature searches.BMC Bioinformatics. 2005 Jan 20;6:12. doi: 10.1186/1471-2105-6-12. BMC Bioinformatics. 2005. PMID: 15661078 Free PMC article.
-
Interoperability with Moby 1.0--it's better than sharing your toothbrush!Brief Bioinform. 2008 May;9(3):220-31. doi: 10.1093/bib/bbn003. Epub 2008 Jan 31. Brief Bioinform. 2008. PMID: 18238804 Review.
-
Representing ontogeny through ontology: a developmental biologist's guide to the gene ontology.Mol Reprod Dev. 2010 Apr;77(4):314-29. doi: 10.1002/mrd.21130. Mol Reprod Dev. 2010. PMID: 19921742 Free PMC article. Review.
Cited by
-
The natural history of molecular functions inferred from an extensive phylogenomic analysis of gene ontology data.PLoS One. 2017 May 3;12(5):e0176129. doi: 10.1371/journal.pone.0176129. eCollection 2017. PLoS One. 2017. PMID: 28467492 Free PMC article.
-
Meta-analysis of nasopharyngeal carcinoma microarray data explores mechanism of EBV-regulated neoplastic transformation.BMC Genomics. 2008 Jul 7;9:322. doi: 10.1186/1471-2164-9-322. BMC Genomics. 2008. PMID: 18605998 Free PMC article.
-
The Compressed Vocabulary of Microbial Life.Front Microbiol. 2021 Jul 7;12:655990. doi: 10.3389/fmicb.2021.655990. eCollection 2021. Front Microbiol. 2021. PMID: 34305827 Free PMC article.
-
GOFFA: gene ontology for functional analysis--a FDA gene ontology tool for analysis of genomic and proteomic data.BMC Bioinformatics. 2006 Sep 6;7 Suppl 2(Suppl 2):S23. doi: 10.1186/1471-2105-7-S2-S23. BMC Bioinformatics. 2006. PMID: 17118145 Free PMC article.
-
Towards refactoring the Molecular Function Ontology with a UML profile for function modeling.J Biomed Semantics. 2017 Oct 4;8(1):48. doi: 10.1186/s13326-017-0152-y. J Biomed Semantics. 2017. PMID: 28978337 Free PMC article.
References
-
- Gene Ontology Consortium http://www.geneontology.org/
-
- Gene Ontology Annotation http://www.ebi.ac.uk/goa
-
- Dwight SS, Harris MA, Dolinski K, Ball CA, Binkley G, Christie KR, Fisk DG, Issel-Tarver L, Schroeder M, Sherlock G, Sethuraman A, Weng S, Botstein D, Cherry JM. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO) Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

