Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast
- PMID: 15889146
- PMCID: PMC1142605
- DOI: 10.1038/sj.emboj.7600670
Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast
Abstract
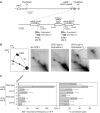
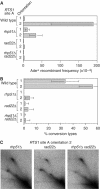
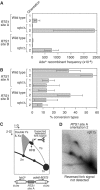
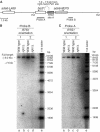
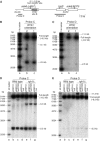
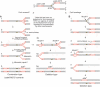
Homologous recombination is believed to play important roles in processing stalled/blocked replication forks in eukaryotes. In accordance with this, recombination is induced by replication fork barriers (RFBs) within the rDNA locus. However, the rDNA locus is a specialised region of the genome, and therefore the action of recombinases at its RFBs may be atypical. We show here for the first time that direct repeat recombination, dependent on Rad22 and Rhp51, is induced by replication fork blockage at a site-specific RFB (RTS1) within a 'typical' genomic locus in fission yeast. Importantly, when the RFB is positioned between the direct repeat, conservative gene conversion events predominate over deletion events. This is consistent with recombination occurring without breakage of the blocked fork. In the absence of the RecQ family DNA helicase Rqh1, deletion events increase dramatically, which correlates with the detection of one-sided DNA double-strand breaks at or near RTS1. These data indicate that Rqh1 acts to prevent blocked replication forks from collapsing and thereby inducing deletion events.
Figures

References
-
- Brewer BJ, Fangman WL (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 - PubMed
-
- Brewer BJ, Fangman WL (1988) A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643 - PubMed
-
- Burkhalter MD, Sogo JM (2004) rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol Cell 15: 409–421 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

