Human T-cell leukemia virus type 1 molecular variants, Vanuatu, Melanesia
- PMID: 15890124
- PMCID: PMC3320372
- DOI: 10.3201/eid1105.041015
Human T-cell leukemia virus type 1 molecular variants, Vanuatu, Melanesia
Abstract
Four of 391 Ni-Vanuatu women were infected with variants of human T-cell leukemia virus type 1 (HTLV-1) Melanesian subtype C. These strains had env nucleotide sequences approximately 99% similar to each other and diverging from the main molecular subtypes of HTLV-1 by 6% to 9%. These strains were likely introduced during ancient human population movements in Melanesia.
Figures
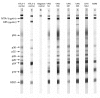
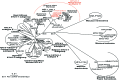

References
-
- Slattery JP, Franchini G, Gessain A. Genomic evolution, patterns of global dissemination, and interspecies transmission of human and simian T-cell leukemia/lymphotropic viruses. Genome Res. 1999;9:525–40. - PubMed
-
- Gessain A, Boeri E, Yanagihara R, Gallo RC, Franchini G. Complete nucleotide sequence of a highly divergent human T-cell leukemia (lymphotropic) virus type I (HTLV-I) variant from Melanesia: genetic and phylogenetic relationship to HTLV-I strains from other geographical regions. J Virol. 1993;67:1015–23. - PMC - PubMed
-
- Gessain A, Yanagihara R, Franchini G, Garruto RM, Jenkins CL, Ajdukiewicz AB, et al. Highly divergent molecular variants of human T-lymphotropic virus type I from isolated populations in Papua New Guinea and the Solomon Islands. Proc Natl Acad Sci U S A. 1991;88:7694–8. 10.1073/pnas.88.17.7694 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
