Coronavirus phylogeny based on a geometric approach
- PMID: 15890535
- PMCID: PMC7111192
- DOI: 10.1016/j.ympev.2005.03.030
Coronavirus phylogeny based on a geometric approach
Abstract
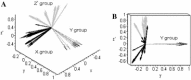
A novel coronavirus has been identified as the cause of the outbreak of severe acute respiratory syndrome (SARS). Previous phylogenetic analyses based on sequence alignments show that SARS-CoVs form a new group distantly related to the other three groups of previously characterized coronaviruses. In this paper, a geometric approach based on the Z-curve representation of the whole genome sequence is proposed to analyze the phylogenetic relationships of coronaviruses. The evolutionary distances are obtained through measuring the differences among the three-dimensional Z-curves. The Z-curve is approximately described by its geometric center and the associated three eigenvectors, which indicate the center position and the trend of the Z-curve, respectively. Although some information is lost due to the approximate description of the Z-curve, the phylogenetic tree constructed based on these parameters is consistent with those of previous analyses. The present method has the merits of simplicity and intuitiveness, but it is still in its premature stage. Because the phylogenetic relationships are inferred from the whole genome, instead of some individual genes, the present method represents a new direction of phylogeny study in the post-genome era.
Figures


References
-
- Cork D.J. Achieving consensus of long genomic sequences with the W-curve. In: Lapointe F., McMorris F.R., Janowitz M., editors. Bioconsensus. vol. 61. American Mathematical Society; Providence, RI: 2003. pp. 123–134. (DIMACS series in discrete mathematics and theoretical computer science).
-
- Cork, D.J., Hutch, T.B., Marland, E., Zmuda, J., 2002. Achieving congruency of phylogenetic trees generated by W-curves of genomic sequences. In: Valafar, F. (Ed.), Techniques in Bioinformatics and Medical Informatics. Ann. N. Y. Acad. Sci. 980, 23–31 - PubMed
-
- Cork, D.J., Toguem, A., 2002. Using fuzzy logic to confirm the integrity of a pattern recognition algorithm for long genomic sequences: the W-curve of genomic sequences. In: Valafar, F. (Ed.), Techniques in Bioinformatics and Medical Informatics. Ann. N. Y. Acad. Sci. 980, 32–40 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

