Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins
- PMID: 15890904
- PMCID: PMC1112111
- DOI: 10.1128/JVI.79.11.6664-6673.2005
Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins
Abstract
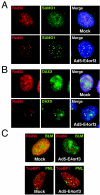
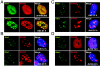
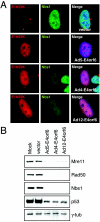
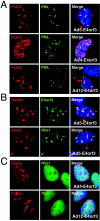
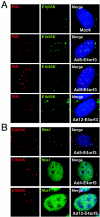
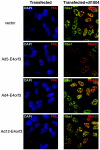
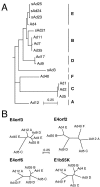
The early transcriptional region 4 (E4) of adenovirus type 5 (Ad5) encodes gene products that modulate splicing, apoptosis, transcription, DNA replication, and repair pathways. Viruses lacking both E4orf3 and E4orf6 have a severe replication defect, partially characterized by the formation of genome concatemers. Concatemer formation is dependent upon the cellular Mre11 complex and is prevented by both the E4orf3 and E4orf6 proteins. The Mre11/Rad50/Nbs1 proteins are targeted for proteasome-mediated degradation by the Ad5 viral E1b55K/E4orf6 complex. The expression of Ad5 E4orf3 causes a redistribution of Mre11 complex members and results in their exclusion from viral replication centers. For this study, we further analyzed the interactions of E4 proteins from different adenovirus serotypes with the Mre11 complex. Analyses of infections with serotypes Ad4 and Ad12 demonstrated that the degradation of Mre11/Rad50/Nbs1 proteins is a conserved feature of the E1b55K/E4orf6 complex. Surprisingly, Nbs1 and Rad50 were localized to the replication centers of both Ad4 and Ad12 viruses prior to Mre11 complex degradation. The transfection of expression vectors for the E4orf3 proteins of Ad4 and Ad12 did not alter the localization of Mre11 complex members. The E4orf3 proteins of Ad4 and Ad12 also failed to complement defects in both concatemer formation and late protein production of a virus with a deletion of E4. These results reveal surprising differences among the highly conserved E4orf3 proteins from different serotypes in the ability to disrupt the Mre11 complex.
Figures

References
-
- Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. - PubMed
-
- Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

