Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome
- PMID: 15892868
- PMCID: PMC1175952
- DOI: 10.1186/gb-2005-6-5-r40
Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome
Abstract
Background: Extensive protein interaction maps are being constructed for yeast, worm, and fly to ask how the proteins organize into pathways and systems, but no such genome-wide interaction map yet exists for the set of human proteins. To prepare for studies in humans, we wished to establish tests for the accuracy of future interaction assays and to consolidate the known interactions among human proteins.
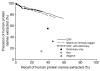
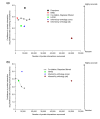
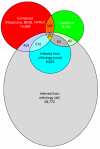
Results: We established two tests of the accuracy of human protein interaction datasets and measured the relative accuracy of the available data. We then developed and applied natural language processing and literature-mining algorithms to recover from Medline abstracts 6,580 interactions among 3,737 human proteins. A three-part algorithm was used: first, human protein names were identified in Medline abstracts using a discriminator based on conditional random fields, then interactions were identified by the co-occurrence of protein names across the set of Medline abstracts, filtering the interactions with a Bayesian classifier to enrich for legitimate physical interactions. These mined interactions were combined with existing interaction data to obtain a network of 31,609 interactions among 7,748 human proteins, accurate to the same degree as the existing datasets.
Conclusion: These interactions and the accuracy benchmarks will aid interpretation of current functional genomics data and provide a basis for determining the quality of future large-scale human protein interaction assays. Projecting from the approximately 15 interactions per protein in the best-sampled interaction set to the estimated 25,000 human genes implies more than 375,000 interactions in the complete human protein interaction network. This set therefore represents no more than 10% of the complete network.
Figures


Similar articles
-
Comparison of human protein-protein interaction maps.Bioinformatics. 2007 Mar 1;23(5):605-11. doi: 10.1093/bioinformatics/btl683. Epub 2007 Jan 19. Bioinformatics. 2007. PMID: 17237052
-
Computational detection of protein complexes in AP-MS experiments.Proteomics. 2012 May;12(10):1663-8. doi: 10.1002/pmic.201100508. Proteomics. 2012. PMID: 22711593 Review.
-
Probabilistic prediction and ranking of human protein-protein interactions.BMC Bioinformatics. 2007 Jul 5;8:239. doi: 10.1186/1471-2105-8-239. BMC Bioinformatics. 2007. PMID: 17615067 Free PMC article.
-
A mouse protein interactome through combined literature mining with multiple sources of interaction evidence.Amino Acids. 2010 Apr;38(4):1237-52. doi: 10.1007/s00726-009-0335-7. Epub 2009 Aug 8. Amino Acids. 2010. PMID: 19669079
-
The Cartographers toolbox: building bigger and better human protein interaction networks.Brief Funct Genomic Proteomic. 2009 Jan;8(1):1-11. doi: 10.1093/bfgp/elp003. Epub 2009 Mar 12. Brief Funct Genomic Proteomic. 2009. PMID: 19282470 Review.
Cited by
-
Extracting relations from traditional Chinese medicine literature via heterogeneous entity networks.J Am Med Inform Assoc. 2016 Mar;23(2):356-65. doi: 10.1093/jamia/ocv092. Epub 2015 Jul 29. J Am Med Inform Assoc. 2016. PMID: 26224335 Free PMC article.
-
IntNetDB v1.0: an integrated protein-protein interaction network database generated by a probabilistic model.BMC Bioinformatics. 2006 Nov 18;7:508. doi: 10.1186/1471-2105-7-508. BMC Bioinformatics. 2006. PMID: 17112386 Free PMC article.
-
Identification of mitochondrial disease genes through integrative analysis of multiple datasets.Methods. 2008 Dec;46(4):248-55. doi: 10.1016/j.ymeth.2008.10.002. Epub 2008 Oct 16. Methods. 2008. PMID: 18930150 Free PMC article.
-
Reconstruction of human protein interolog network using evolutionary conserved network.BMC Bioinformatics. 2007 May 10;8:152. doi: 10.1186/1471-2105-8-152. BMC Bioinformatics. 2007. PMID: 17493278 Free PMC article.
-
Viral organization of human proteins.PLoS One. 2010 Aug 25;5(8):e11796. doi: 10.1371/journal.pone.0011796. PLoS One. 2010. PMID: 20827298 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

