Dissection of a DNA-damage-induced transcriptional network using a combination of microarrays, RNA interference and computational promoter analysis
- PMID: 15892871
- PMCID: PMC1175955
- DOI: 10.1186/gb-2005-6-5-r43
Dissection of a DNA-damage-induced transcriptional network using a combination of microarrays, RNA interference and computational promoter analysis
Abstract
Background: Gene-expression microarrays and RNA interferences (RNAi) are among the most prominent techniques in functional genomics. The combination of the two holds promise for systematic, large-scale dissection of transcriptional networks. Recent studies, however, raise the concern that nonspecific responses to small interfering RNAs (siRNAs) might obscure the consequences of silencing the gene of interest, throwing into question the ability of this experimental strategy to achieve precise network dissections.
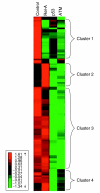
Results: We used microarrays and RNAi to dissect a transcriptional network induced by DNA damage in a human cellular system. We recorded expression profiles with and without exposure of the cells to a radiomimetic drug that induces DNA double-strand breaks (DSBs). Profiles were measured in control cells and in cells knocked-down for the Rel-A subunit of NFkappaB and for p53, two pivotal stress-induced transcription factors, and for the protein kinase ATM, the major transducer of the cellular responses to DSBs. We observed that NFkappaB and p53 mediated most of the damage-induced gene activation; that they controlled the activation of largely disjoint sets of genes; and that ATM was required for the activation of both pathways. Applying computational promoter analysis, we demonstrated that the dissection of the network into ATM/NFkappaB and ATM/p53-mediated arms was highly accurate.
Conclusions: Our results demonstrate that the combined experimental strategy of expression arrays and RNAi is indeed a powerful method for the dissection of complex transcriptional networks, and that computational promoter analysis can provide a strong complementary means for assessing the accuracy of this dissection.
Figures
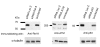
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

