Expanding the frontiers of existing antiviral drugs: possible effects of HIV-1 protease inhibitors against SARS and avian influenza
- PMID: 15893956
- PMCID: PMC7108403
- DOI: 10.1016/j.jcv.2005.03.005
Expanding the frontiers of existing antiviral drugs: possible effects of HIV-1 protease inhibitors against SARS and avian influenza
Abstract
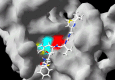
When unexpected diseases such as the severe acute respiratory syndrome (SARS) and avian influenza become a serious threat to public health, an immediate response is imperative. This should take into consideration existing licensed antiviral drugs against other viral diseases already known to be safe for use in humans. In this report, evidence is presented that HIV-1 protease inhibitors (PIs) currently used in anti-HIV-1 therapies might exert some effects on SARS and perhaps, on avian influenza. Evidence for the potential benefits of PIs against the SARS coronavirus (SARS-CoV) is provided by empirical clinical studies, in vivo viral inhibition assays and computational simulations of the docking of these compounds to the active site of the main SARS-CoV protease. As suggested by in silico docking of these molecules to a theoretical model of a subunit of type A influenza virus RNA-dependent RNA polymerase, there also exists a remote possibility that these PIs may have an effect on avian influenza viruses. Although this evidence is still far from being definitive, the results so far obtained suggest that PIs should be seriously taken into consideration for further testing as potential therapeutic agents for SARS and avian influenza.
Figures



References
-
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. - PubMed
-
- Cassone A., De Bernardis F., Torosantucci A., Tacconelli E., Tumbarello M., Cauda R. In vitro and in vivo anticandidal activity of human immunodeficiency virus protease inhibitors. J Infect Dis. 1999;180:448–453. - PubMed
-
- Chan K.S., Lai S.T., Chu C.M., Tsui E., Tam C.Y., Wong M.M. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicentre retrospective matched cohort study. Hong Kong Med J. 2003;9:399–406. - PubMed
-
- Chen X.P., Cao Y. Consideration of highly active antiretroviral therapy in the prevention and treatment of severe acute respiratory syndrome. Clin Infect Dis. 2004;38:1030–1032. - PubMed
-
- Chen X.P., Li G.H., Tang X.P., Xiong Y., Chen X.J., Cao Y. Lack of severe acute respiratory syndrome in 19 AIDS patients hospitalised together. J Acquir Immun Defic Syndr. 2003;34:242–243. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

