Molecular dynamics of a protein surface: ion-residues interactions
- PMID: 15894639
- PMCID: PMC1366628
- DOI: 10.1529/biophysj.105.058917
Molecular dynamics of a protein surface: ion-residues interactions
Abstract
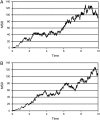
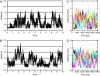
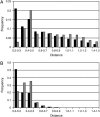
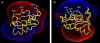
Time-resolved measurements indicated that protons could propagate on the surface of a protein or a membrane by a special mechanism that enhanced the shuttle of the proton toward a specific site. It was proposed that a suitable location of residues on the surface contributes to the proton shuttling function. In this study, this notion was further investigated by the use of molecular dynamics simulations, where Na(+) and Cl(-) are the ions under study, thus avoiding the necessity for quantum mechanical calculations. Molecular dynamics simulations were carried out using as a model a few Na(+) and Cl(-) ions enclosed in a fully hydrated simulation box with a small globular protein (the S6 of the bacterial ribosome). Three independent 10-ns-long simulations indicated that the ions and the protein's surface were in equilibrium, with rapid passage of the ions between the protein's surface and the bulk. However, it was noted that close to some domains the ions extended their duration near the surface, thus suggesting that the local electrostatic potential hindered their diffusion to the bulk. During the time frame in which the ions were detained next to the surface, they could rapidly shuttle between various attractor sites located under the electrostatic umbrella. Statistical analysis of the molecular dynamics and electrostatic potential/entropy consideration indicated that the detainment state is an energetic compromise between attractive forces and entropy of dilution. The similarity between the motion of free ions next to a protein and the proton transfer on the protein's surface are discussed.
Figures
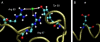
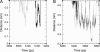
Similar articles
-
Protein surface dynamics: interaction with water and small solutes.J Biol Phys. 2005 Dec;31(3-4):433-52. doi: 10.1007/s10867-005-0171-2. J Biol Phys. 2005. PMID: 23345909 Free PMC article.
-
The dynamics of proton transfer between adjacent sites.Photochem Photobiol Sci. 2006 Jun;5(6):531-7. doi: 10.1039/b515887g. Epub 2006 Mar 3. Photochem Photobiol Sci. 2006. PMID: 16761081
-
Interfacial interactions of glutamate, water and ions with carbon nanopore evaluated by molecular dynamics simulations.Biochim Biophys Acta. 2007 Sep;1768(9):2319-41. doi: 10.1016/j.bbamem.2007.06.006. Epub 2007 Jun 15. Biochim Biophys Acta. 2007. PMID: 17631857
-
Biophysical aspects of intra-protein proton transfer.Biochim Biophys Acta. 2000 May 12;1458(1):120-34. doi: 10.1016/s0005-2728(00)00063-3. Biochim Biophys Acta. 2000. PMID: 10812028 Review.
-
Structure, dynamics and interaction with kinase targets: computer simulations of calmodulin.Biochim Biophys Acta. 2004 Mar 11;1697(1-2):289-300. doi: 10.1016/j.bbapap.2003.11.032. Biochim Biophys Acta. 2004. PMID: 15023369 Review.
Cited by
-
Ion-induced alterations of the local hydration environment elucidate Hofmeister effect in a simple classical model of Trp-cage miniprotein.J Mol Model. 2017 Sep 27;23(10):298. doi: 10.1007/s00894-017-3471-0. J Mol Model. 2017. PMID: 28956172
-
Site Identification by Ligand Competitive Saturation-Biologics Approach for Structure-Based Protein Charge Prediction.Mol Pharm. 2023 May 1;20(5):2600-2611. doi: 10.1021/acs.molpharmaceut.3c00064. Epub 2023 Apr 5. Mol Pharm. 2023. PMID: 37017675 Free PMC article.
-
Polymorphic Protein Crystal Growth: Influence of Hydration and Ions in Glucose Isomerase.Cryst Growth Des. 2014 Jan 2;14(1):46-57. doi: 10.1021/cg401063b. Cryst Growth Des. 2014. PMID: 24955067 Free PMC article.
-
SPEADI: Accelerated Analysis of IDP-Ion Interactions from MD-Trajectories.Biology (Basel). 2023 Apr 10;12(4):581. doi: 10.3390/biology12040581. Biology (Basel). 2023. PMID: 37106781 Free PMC article.
-
The dynamics of Ca2+ ions within the solvation shell of calbindin D9k.PLoS One. 2011 Feb 22;6(2):e14718. doi: 10.1371/journal.pone.0014718. PLoS One. 2011. PMID: 21364983 Free PMC article.
References
-
- Klotz, I. M. 1958. Protein hydration and behavior; many aspects of protein behavior can be interpreted in terms of frozen water of hydration. Science. 128:815–822. - PubMed
-
- Tanford, C. 1969. Extension of the theory of linked functions to incorporate the effects of protein hydration. J. Mol. Biol. 39:539–544. - PubMed
-
- Blicharska, B., Z. Florkowski, J. W. Hennel, G. Held, and F. Noack. 1970. Investigation of protein hydration by proton spin relaxation time measurements. Biochim. Biophys. Acta. 207:381–389. - PubMed
-
- Yang, P. H., and J. A. Rupley. 1979. Protein-water interactions. Heat capacity of the lysozyme-water system. Biochemistry. 18:2654–2661. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

