Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase
- PMID: 15894717
- PMCID: PMC1143070
- DOI: 10.1105/tpc.105.031393
Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase
Abstract
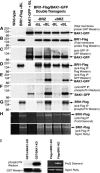
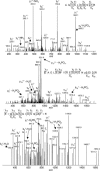
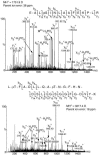
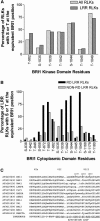
Brassinosteroids (BRs) regulate multiple aspects of plant growth and development and require an active BRASSINOSTEROID-INSENSITIVE1 (BRI1) and BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) for hormone perception and signal transduction. Many animal receptor kinases exhibit ligand-dependent oligomerization followed by autophosphorylation and activation of the intracellular kinase domain. To determine if early events in BR signaling share this mechanism, we used coimmunoprecipitation of epitope-tagged proteins to show that in vivo association of BRI1 and BAK1 was affected by endogenous and exogenous BR levels and that phosphorylation of both BRI1 and BAK1 on Thr residues was BR dependent. Immunoprecipitation of epitope-tagged BRI1 from Arabidopsis thaliana followed by liquid chromatography-tandem mass spectrometry (LC/MS/MS) identified S-838, S-858, T-872, and T-880 in the juxtamembrane region, T-982 in the kinase domain, and S-1168 in C-terminal region as in vivo phosphorylation sites of BRI1. MS analysis also strongly suggested that an additional two residues in the juxtamembrane region and three sites in the activation loop of kinase subdomain VII/VIII were phosphorylated in vivo. We also identified four specific BAK1 autophosphorylation sites in vitro using LC/MS/MS. Site-directed mutagenesis of identified and predicted BRI1 phosphorylation sites revealed that the highly conserved activation loop residue T-1049 and either S-1044 or T-1045 were essential for kinase function in vitro and normal BRI1 signaling in planta. Mutations in the juxtamembrane or C-terminal regions had only small observable effects on autophosphorylation and in planta signaling but dramatically affected phosphorylation of a peptide substrate in vitro. These findings are consistent with many aspects of the animal receptor kinase model in which ligand-dependent autophosphorylation of the activation loop generates a functional kinase, whereas phosphorylation of noncatalytic intracellular domains is required for recognition and/or phosphorylation of downstream substrates.
Figures




References
-
- Adams, J.A. (2003). Activation loop phosphorylation and catalysis in protein kinases: Is there functional evidence for the autoinhibitor model? Biochemistry 42, 601–607. - PubMed
-
- Altmann, T. (1999). Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta 208, 1–11. - PubMed
-
- Baxter, R.M., Secrist, J.P., Vaillancourt, R.R., and Kazlauskas, A. (1998). Full activation of the platelet-derived growth factor beta-receptor kinase involves multiple events. J. Biol. Chem. 273, 17050–17055. - PubMed
-
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20, 1195–1197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

