Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels
- PMID: 15897982
- PMCID: PMC1226195
- DOI: 10.1086/431366
Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels
Erratum in
- Am J Hum Genet. 2008 Jan;82(1):251
Abstract
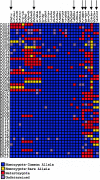
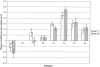
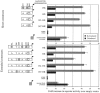
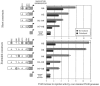
Elevated plasma levels of C-reactive protein (CRP), an inflammation-sensitive marker, have emerged as an important predictor of future cardiovascular disease and metabolic abnormalities in apparently healthy men and women. Here, we performed a systematic survey of common nucleotide variation across the genomic region encompassing the CRP gene locus. Of the common single-nucleotide polymorphisms (SNPs) identified, several in the CRP promoter region are strongly associated with CRP levels in a large cohort study of cardiovascular risk in European American and African American young adults. We also demonstrate the functional importance of these SNPs in vitro.
Figures



References
Web Resources
-
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/index.html (for tagSNPs 790 [rs3093058], 1440 [rs3091244], 1919 [rs1417938], 2667 [rs1800947], 3006 [rs3093066], 3872 [rs1205], and 5237 [rs2808630])
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the CRP gene [accession number AF449713])
-
- SeattleSNPs Program for Genomic Applications, http://pga.gs.washington.edu/protocols/dnapanel_protocol.html
References
-
- Abernathy TJ, Avery OT (1941) The occurrence during acute infections of a protein not normally present in the blood. I. Distribution of the reactive protein in patient’s sera and the effect of calcium on the flocculation reaction with C-polysaccharide of pneumococcus. J Exp Med 73:173–182 - PMC - PubMed
-
- Albert MA, Glynn RJ, Buring J, Ridker PM (2004) C-reactive protein levels among women of various ethnic groups living in the United States (from the Women’s Health Study). Am J Cardiol 93:1238–1242 - PubMed
-
- Anand SS, Razak F, Yi Q, Davis B, Jacobs R, Vuksan V, Lonn E, Teo K, McQueen M, Yusuf S (2004) C-reactive protein as a screening test for cardiovascular risk in a multiethnic population. Arterioscler Thromb Vasc Biol 24:1509–1515 - PubMed
-
- Brull DJ, Serrano N, Zito F, Jones L, Montgomery HE, Rumley A, Sharma P, Lowe GD, World MJ, Humphries SE, Hingorani AD (2003) Human CRP gene polymorphism influences CRP levels: implications for the prediction and pathogenesis of coronary heart disease. Arterioscler Thromb Vasc Biol 23:2063–2069 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 HL066642/HL/NHLBI NIH HHS/United States
- N01 HC095095/HL/NHLBI NIH HHS/United States
- N01 HC048048/HL/NHLBI NIH HHS/United States
- HL71017/HL/NHLBI NIH HHS/United States
- N01-HC-48047/HC/NHLBI NIH HHS/United States
- N01 HC048049/HL/NHLBI NIH HHS/United States
- N01-HC-95095/HC/NHLBI NIH HHS/United States
- N01-HC-48050/HC/NHLBI NIH HHS/United States
- N01-HC-48049/HC/NHLBI NIH HHS/United States
- N01 HC048047/HL/NHLBI NIH HHS/United States
- N01 HC048050/HL/NHLBI NIH HHS/United States
- HL66682/HL/NHLBI NIH HHS/United States
- N01-HC-48048/HC/NHLBI NIH HHS/United States
- HL66642/HL/NHLBI NIH HHS/United States
- U01 HL066682/HL/NHLBI NIH HHS/United States
- R01 HL071017/HL/NHLBI NIH HHS/United States
- N01-HC-05187/HC/NHLBI NIH HHS/United States
- N01 HC005187/HL/NHLBI NIH HHS/United States
- N01 HC045134/HC/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

