Spatial invasion by a mutant pathogen
- PMID: 15899503
- PMCID: PMC3057370
- DOI: 10.1016/j.jtbi.2005.03.016
Spatial invasion by a mutant pathogen
Abstract
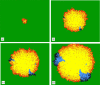
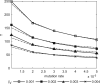
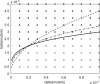
Imagine a pathogen that is spreading radially as a circular wave through a population of susceptible hosts. In the interior of this circular region, the infection dies out due to a subcritical density of susceptibles. If a mutant pathogen, having some advantage over wild-type pathogens, arises in this region it is likely to die out without leaving a noticeable trace. Mutants that arise closer to the infection wavefront have access to more susceptible hosts and thus are more likely to become established and perhaps (locally) out-compete the original pathogen. Among the factors (position, transmission rate, pathogen-induced death rate) that influence the fate of a mutant, which are most important? What does this tell us about the types of mutants that are likely to invade and become established? How do such tendencies serve to steer the evolution of pathogens in a spatial setting? Do different types of models of the same phenomena lead to similar conclusions? We address these issues from the point of view of an individual-based stochastic spatial model of host-pathogen interactions. We consider the probability of a successful invasion by a single mutant as a function of the transmissibility and virulence strengths and the mutant position in the wavefront. Next, for a version of the model in which mutations arise spontaneously, we obtain analytical and simulation results on the mean time to a successful invasion. We also use our model predictions to gain insight into experimental data on bacteriophage plaques. Finally, we compare our results to those based on ordinary and partial differential equations to better understand how different models might influence our predictions on the fate of a mutant pathogen.
Figures









Similar articles
-
Virulence of vector-borne pathogens. A stochastic automata model of perpetuation.Ann N Y Acad Sci. 1994 Dec 15;740:249-59. doi: 10.1111/j.1749-6632.1994.tb19875.x. Ann N Y Acad Sci. 1994. PMID: 7840455 Review.
-
Pathogen transmission at stage-structured infectious patches: Killers and vaccinators.J Theor Biol. 2018 Jan 7;436:51-63. doi: 10.1016/j.jtbi.2017.09.029. Epub 2017 Sep 28. J Theor Biol. 2018. PMID: 28966110
-
The impact of cross-immunity, mutation and stochastic extinction on pathogen diversity.Proc Biol Sci. 2004 Dec 7;271(1556):2431-8. doi: 10.1098/rspb.2004.2877. Proc Biol Sci. 2004. PMID: 15590592 Free PMC article.
-
Stochastic models for competing species with a shared pathogen.Math Biosci Eng. 2012 Jul;9(3):461-85. doi: 10.3934/mbe.2012.9.461. Math Biosci Eng. 2012. PMID: 22881022
-
Processes controlling the transmission of bacterial pathogens in the environment.Res Microbiol. 2007 Apr;158(3):195-202. doi: 10.1016/j.resmic.2006.12.005. Epub 2007 Jan 13. Res Microbiol. 2007. PMID: 17350808 Review.
Cited by
-
Fitness benefits of low infectivity in a spatially structured population of bacteriophages.Proc Biol Sci. 2013 Nov 13;281(1774):20132563. doi: 10.1098/rspb.2013.2563. Print 2014 Jan 7. Proc Biol Sci. 2013. PMID: 24225463 Free PMC article.
-
Quantitative understanding of cell signaling: the importance of membrane organization.Curr Opin Biotechnol. 2010 Oct;21(5):677-82. doi: 10.1016/j.copbio.2010.08.006. Epub 2010 Sep 9. Curr Opin Biotechnol. 2010. PMID: 20829029 Free PMC article. Review.
-
The impact of spatial structure on viral genomic diversity generated during adaptation to thermal stress.PLoS One. 2014 Feb 12;9(2):e88702. doi: 10.1371/journal.pone.0088702. eCollection 2014. PLoS One. 2014. PMID: 24533140 Free PMC article.
-
Modelling the spatial dynamics of plasmid transfer and persistence.Microbiology (Reading). 2007 Aug;153(Pt 8):2803-2816. doi: 10.1099/mic.0.2006/004531-0. Microbiology (Reading). 2007. PMID: 17660444 Free PMC article.
-
Bacteriophage secondary infection.Virol Sin. 2015 Feb;30(1):3-10. doi: 10.1007/s12250-014-3547-2. Epub 2015 Jan 13. Virol Sin. 2015. PMID: 25595214 Free PMC article. Review.
References
-
- Boots M, Hudson PJ, Sasaki A. Large shifts in pathogen virulence relate to host population structure. Science. 2004;303:842–844. - PubMed
-
- Bremermann HJ, Thieme HR. A competitive-exclusion principle for pathogen virulence. J. Math. Biol. 1989;27:179–190. - PubMed
-
- Durrett R. Lecture Notes on Particle Systems and Percolation. Wadsworth; Belmont, CA: 1988.
-
- Durrett R, Neuhauser C. Particle systems and reaction diffusion equations. Ann. Probab. 1994;22:289–333.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

