Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA
- PMID: 15899842
- PMCID: PMC1140639
- DOI: 10.1128/MCB.25.11.4359-4370.2005
Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA
Abstract
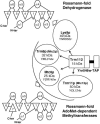
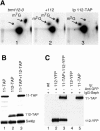
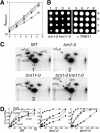
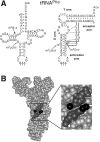
N(2)-Monomethylguanosine-10 (m(2)G10) and N(2),N(2)-dimethylguanosine-26 (m(2)(2)G26) are the only two guanosine modifications that have been detected in tRNA from nearly all archaea and eukaryotes but not in bacteria. In Saccharomyces cerevisiae, formation of m(2)(2)G26 is catalyzed by Trm1p, and we report here the identification of the enzymatic activity that catalyzes the formation of m(2)G10 in yeast tRNA. It is composed of at least two subunits that are associated in vivo: Trm11p (Yol124c), which is the catalytic subunit, and Trm112p (Ynr046w), a putative zinc-binding protein. While deletion of TRM11 has no detectable phenotype under laboratory conditions, deletion of TRM112 leads to a severe growth defect, suggesting that it has additional functions in the cell. Indeed, Trm112p is associated with at least four proteins: two tRNA methyltransferases (Trm9p and Trm11p), one putative protein methyltransferase (Mtc6p/Ydr140w), and one protein with a Rossmann fold dehydrogenase domain (Lys9p/Ynr050c). In addition, TRM11 interacts genetically with TRM1, thus suggesting that the absence of m(2)G10 and m(2)(2)G26 affects tRNA metabolism or functioning.
Figures



References
-
- Aravind, L., and E. V. Koonin. 2001. THUMP: a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 26:215-217. - PubMed
-
- Arluison, V., C. Hountondji, B. Robert, and H. Grosjean. 1998. Transfer RNA-pseudouridine synthetase Pus1 of Saccharomyces cerevisiae contains one atom of zinc essential for its native conformation and tRNA recognition. Biochemistry 37:7268-7276. - PubMed
-
- Armengaud, J., J. Urbonavicius, B. Fernandez, G. Chaussinand, J. M. Bujnicki, and H. Grosjean. 2004. N2-Methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J. Biol. Chem. 279:37142-37152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
