Dynamics of transcription and mRNA export
- PMID: 15901505
- PMCID: PMC4943869
- DOI: 10.1016/j.ceb.2005.04.004
Dynamics of transcription and mRNA export
Abstract
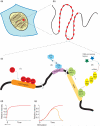
Understanding the different molecular mechanisms responsible for gene expression has been a central interest of molecular biologists for several decades. Transcription, the initial step of gene expression, consists of converting the genetic code into a dynamic messenger RNA that will specify a required cellular function following translocation to the cytoplasm and translation. We now possess an in-depth understanding of the mechanism and regulations of transcription. By contrast, an understanding of the dynamics of an individual gene's expression in real time is just beginning to emerge following recent technological developments.
Figures
References
-
- Cramer P. RNA polymerase II structure: from core to functional complexes. Curr Opin Genet Dev. 2004;14:218–226. - PubMed
-
- Fahrenkrog B, Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. - PubMed
-
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. - PubMed
-
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

