Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C
- PMID: 15901673
- PMCID: PMC1142559
- DOI: 10.1101/gad.1305005
Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C
Abstract
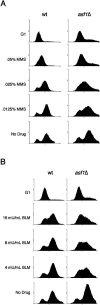
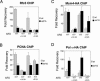
Chromatin assembly and DNA replication are temporally coupled, and DNA replication in the absence of histone synthesis causes inviability. Here we demonstrate that chromatin assembly factor Asf1 also affects DNA replication. In budding yeast cells lacking Asf1, the amounts of several DNA replication proteins, including replication factor C (RFC), proliferating cell nuclear antigen (PCNA), and DNA polymerase epsilon (Pol epsilon), are reduced at stalled replication forks. In contrast, DNA polymerase alpha (Pol alpha) accumulates to higher than normal levels at stalled forks in asf1Delta cells. Using purified, recombinant proteins, we demonstrate that RFC directly binds Asf1 and can recruit Asf1 to DNA molecules in vitro. We conclude that histone chaperone protein Asf1 maintains a subset of replication elongation factors at stalled replication forks and directly interacts with the replication machinery.
Figures





References
-
- Bell S.P. and Dutta, A. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71: 333-374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
