Role of pi dimers in coupling ("handcuffing") of plasmid R6K's gamma ori iterons
- PMID: 15901701
- PMCID: PMC1112066
- DOI: 10.1128/JB.187.11.3779-3785.2005
Role of pi dimers in coupling ("handcuffing") of plasmid R6K's gamma ori iterons
Abstract
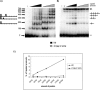
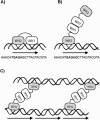
One proposed mechanism of replication inhibition in iteron-containing plasmids (ICPs) is "handcuffing," in which the coupling of origins via iteron-bound replication initiator (Rep) protein turns off origin function. In minimal R6K replicons, copy number control requires the interaction of plasmid-encoded pi protein with the seven 22-bp iterons of the gamma origin of replication. Like other related Rep proteins, pi exists as both monomers and dimers. However, the ability of pi dimers to bind iterons distinguishes R6K from most other ICPs, where only monomers have been observed to bind iterons. Here, we describe experiments to determine if monomers or dimers of pi protein are involved in the formation of handcuffed complexes. Standard ligation enhancement assays were done using pi variants with different propensities to bind iterons as monomers or dimers. Consistent with observations from several ICPs, a hyperreplicative variant (pi.P106L(wedge)F107S) exhibits deficiencies in handcuffing. Additionally, a novel dimer-biased variant of pi protein (pi.M36A(wedge)M38A), which lacks initiator function, handcuffs iteron-containing DNA more efficiently than does wild-type pi. The data suggest that pi dimers mediate handcuffing, supporting our previously proposed model of handcuffing in the gamma ori system. Thus, dimers of pi appear to possess three distinct inhibitory functions with respect to R6K replication: transcriptional autorepression of pi expression, in cis competition (for origin binding) with monomeric activator pi, and handcuffing-mediated inhibition of replication in trans.
Figures



References
-
- Abhyankar, M. M., J. M. Reddy, R. Sharma, E. Bullesbach, and D. Bastia. 2004. Biochemical investigations of control of replication initiation of plasmid R6K. J. Biol. Chem. 279:6711-6719. - PubMed
-
- Chattoraj, D. K., and T. D. Schneider. 1997. Replication control of plasmid P1 and its host chromosome: the common ground. Prog. Nucleic Acid Res. Mol. Biol. 57:145-186. - PubMed
-
- Das, N., and D. K. Chattoraj. 2004. Origin pairing (‘handcuffing’) and unpairing in the control of P1 plasmid replication. Mol. Microbiol. 54:836-849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

