Prediction of functional modules based on comparative genome analysis and Gene Ontology application
- PMID: 15901854
- PMCID: PMC1130488
- DOI: 10.1093/nar/gki573
Prediction of functional modules based on comparative genome analysis and Gene Ontology application
Abstract
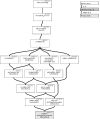
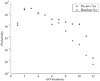
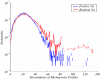
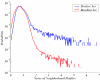
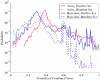
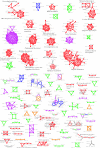
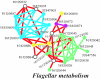
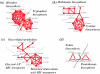
We present a computational method for the prediction of functional modules encoded in microbial genomes. In this work, we have also developed a formal measure to quantify the degree of consistency between the predicted and the known modules, and have carried out statistical significance analysis of consistency measures. We first evaluate the functional relationship between two genes from three different perspectives--phylogenetic profile analysis, gene neighborhood analysis and Gene Ontology assignments. We then combine the three different sources of information in the framework of Bayesian inference, and we use the combined information to measure the strength of gene functional relationship. Finally, we apply a threshold-based method to predict functional modules. By applying this method to Escherichia coli K12, we have predicted 185 functional modules. Our predictions are highly consistent with the previously known functional modules in E.coli. The application results have demonstrated that our approach is highly promising for the prediction of functional modules encoded in a microbial genome.
Figures

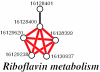
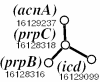
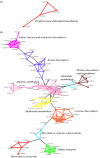

Similar articles
-
Prediction of functional modules based on gene distributions in microbial genomes.Genome Inform. 2005;16(2):247-59. Genome Inform. 2005. PMID: 16901107
-
Gene fusions and gene duplications: relevance to genomic annotation and functional analysis.BMC Genomics. 2005 Mar 9;6:33. doi: 10.1186/1471-2164-6-33. BMC Genomics. 2005. PMID: 15757509 Free PMC article.
-
[Comparison of algorithms for prediction of related proteins using the method of phylogenetic profiles].Biomed Khim. 2009 Sep-Oct;55(5):534-43. Biomed Khim. 2009. PMID: 20017387 Russian.
-
Ontology-driven approaches to analyzing data in functional genomics.Methods Mol Biol. 2006;316:67-86. doi: 10.1385/1-59259-964-8:67. Methods Mol Biol. 2006. PMID: 16671401 Review.
-
Function prediction and protein networks.Curr Opin Cell Biol. 2003 Apr;15(2):191-8. doi: 10.1016/s0955-0674(03)00009-7. Curr Opin Cell Biol. 2003. PMID: 12648675 Review.
Cited by
-
Hierarchical classification of functionally equivalent genes in prokaryotes.Nucleic Acids Res. 2007;35(7):2125-40. doi: 10.1093/nar/gkl1114. Epub 2007 Mar 11. Nucleic Acids Res. 2007. PMID: 17353185 Free PMC article.
-
Transcriptional regulatory network refinement and quantification through kinetic modeling, gene expression microarray data and information theory.BMC Bioinformatics. 2007 Jan 23;8:20. doi: 10.1186/1471-2105-8-20. BMC Bioinformatics. 2007. PMID: 17244365 Free PMC article.
-
GO-based functional dissimilarity of gene sets.BMC Bioinformatics. 2011 Sep 1;12:360. doi: 10.1186/1471-2105-12-360. BMC Bioinformatics. 2011. PMID: 21884611 Free PMC article.
-
From disease ontology to disease-ontology lite: statistical methods to adapt a general-purpose ontology for the test of gene-ontology associations.Bioinformatics. 2009 Jun 15;25(12):i63-8. doi: 10.1093/bioinformatics/btp193. Bioinformatics. 2009. PMID: 19478018 Free PMC article.
-
UFO: A tool for unifying biomedical ontology-based semantic similarity calculation, enrichment analysis and visualization.PLoS One. 2020 Jul 9;15(7):e0235670. doi: 10.1371/journal.pone.0235670. eCollection 2020. PLoS One. 2020. PMID: 32645039 Free PMC article.
References
-
- Kremling A., Jahreis K., Lengeler J.W., Gilles E.D. The organization of metabolic reaction networks: a signal-oriented approach to cellular models. Metab. Eng. 2000;2:190–200. - PubMed
-
- Wagner R. Transcription Regulation in Prokaryotes. Oxford, UK: Oxford University Press; 2000.
-
- Stephanopoulos G.N., Aristidou A.A., Nielsen J. Metabolic Engineering: Principles and Methodologies. San Diego, CA: Academic Press; 1998.
-
- Zhou J., Thompson D.K., Xu Y., Tiedje J.M. Microbial Functional Genomics. Hoboken, NJ: John Wiley & Sons, Inc.; 2004.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

