Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase
- PMID: 15902263
- PMCID: PMC1440517
- DOI: 10.1038/nature03580
Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase
Abstract
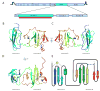
Hepatitis C virus (HCV) is a human pathogen affecting nearly 3% of the world's population. Chronic infections can lead to cirrhosis and liver cancer. The RNA replication machine of HCV is a multi-subunit membrane-associated complex. The non-structural protein NS5A is an active component of HCV replicase, as well as a pivotal regulator of replication and a modulator of cellular processes ranging from innate immunity to dysregulated cell growth. NS5A is a large phosphoprotein (56-58 kDa) with an amphipathic alpha-helix at its amino terminus that promotes membrane association. After this helix region, NS5A is organized into three domains. The N-terminal domain (domain I) coordinates a single zinc atom per protein molecule. Mutations disrupting either the membrane anchor or zinc binding of NS5A are lethal for RNA replication. However, probing the role of NS5A in replication has been hampered by a lack of structural information about this multifunctional protein. Here we report the structure of NS5A domain I at 2.5-A resolution, which contains a novel fold, a new zinc-coordination motif and a disulphide bond. We use molecular surface analysis to suggest the location of protein-, RNA- and membrane-interaction sites.
Conflict of interest statement
Figures



References
-
- Anonymous World Health Organization - Hepatitis C: global prevalence. Wkly Epidemiol Rec. 1997;72:341–4. - PubMed
-
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. - PubMed
-
- Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. - PubMed
-
- Tellinghuisen TL, Rice CM. Interaction between hepatitis C virus proteins and host cell factors. Current Opinion in Microbiology. 2002;5:419–27. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

