Genetic variation in the human androgen receptor gene is the major determinant of common early-onset androgenetic alopecia
- PMID: 15902657
- PMCID: PMC1226186
- DOI: 10.1086/431425
Genetic variation in the human androgen receptor gene is the major determinant of common early-onset androgenetic alopecia
Abstract
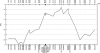
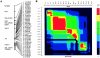
Androgenetic alopecia (AGA), or male-pattern baldness, is the most common form of hair loss. Its pathogenesis is androgen dependent, and genetic predisposition is the major requirement for the phenotype. We demonstrate that genetic variability in the androgen receptor gene (AR) is the cardinal prerequisite for the development of early-onset AGA, with an etiological fraction of 0.46. The investigation of a large number of genetic variants covering the AR locus suggests that a polyglycine-encoding GGN repeat in exon 1 is a plausible candidate for conferring the functional effect. The X-chromosomal location of AR stresses the importance of the maternal line in the inheritance of AGA.
Figures
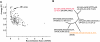
References
Web Resources
-
- Ensembl, http://www.ensembl.org/ (for AR locus information)
-
- HapMap, http://www.hapmap.org/ (for pairwise LD on the X chromosome [genotypes queried in November 2004])
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AGA and AR) - PubMed
-
- University of California–Santa Cruz (UCSC) Genome Bioinformatics, http://genome.ucsc.edu/ (for X-chromosomal recombination rates and definition of the reference strand for SNP allele calling)
References
-
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 - PubMed
-
- Bamshad M, Wooding SP (2003) Signatures of natural selection in the human genome. Nat Rev Genet 4:99–111 - PubMed
-
- ——— (2004b) Maximum-likelihood estimation of haplotype frequencies in nuclear families. Genet Epidemiol 27:21–32 - PubMed
-
- Beilin J, Ball EM, Favaloro JM, Zajac JD (2000) Effect of the androgen receptor CAG repeat polymorphism on transcriptional activity: specificity in prostate and non-prostate cell lines. J Mol Endocrinol 25:85–96 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

