Characterization of subsets of CD4+ memory T cells reveals early branched pathways of T cell differentiation in humans
- PMID: 15905333
- PMCID: PMC1131816
- DOI: 10.1073/pnas.0409720102
Characterization of subsets of CD4+ memory T cells reveals early branched pathways of T cell differentiation in humans
Abstract
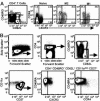
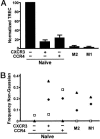
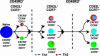
The pathways for differentiation of human CD4(+) T cells into functionally distinct subsets of memory cells in vivo are unknown. The identification of these subsets and pathways has clear implications for the design of vaccines and immune-targeted therapies. Here, we show that populations of apparently naive CD4(+) T cells express the chemokine receptors CXCR3 or CCR4 and demonstrate patterns of gene expression and functional responses characteristic of memory cells. The proliferation history and T cell receptor repertoire of these chemokine-receptor(+) cells suggest that they are very early memory CD4(+) T cells that have "rested down" before acquiring the phenotypes described for "central" or "effector" memory T cells. In addition, the chemokine-receptor(+) "naive" populations contain Th1 and Th2 cells, respectively, demonstrating that Th1/Th2 differentiation can occur very early in vivo in the absence of markers conventionally associated with memory cells. We localized ligands for CXCR3 and CCR4 to separate foci in T cell zones of tonsil, suggesting that the chemokine-receptor(+) subsets may be recruited and contribute to segregated, polarized microenvironments within lymphoid organs. Importantly, our data suggest that CD4(+) T cells do not differentiate according to a simple schema from naive --> CD45RO(+) noneffector/central memory --> effector/effector memory cells. Rather, developmental pathways branch early on to yield effector/memory populations that are highly heterogeneous and multifunctional and have the potential to become stable resting cells.
Figures


References
-
- Kaech, S. M., Wherry, E. J. & Ahmed, R. (2002) Nat. Rev. Immunol. 2, 251–262. - PubMed
-
- Seder, R. A. & Ahmed, R. (2003) Nat. Immunol. 4, 835–842. - PubMed
-
- Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. (1999) Nature 401, 708–712. - PubMed
-
- Sallusto, F., Geginat, J. & Lanzavecchia, A. (2004) Annu. Rev. Immunol. 22, 745–763. - PubMed
-
- Yamamoto, J., Adachi, Y., Onoue, Y., Adachi, Y. S., Okabe, Y., Itazawa, T., Toyoda, M., Seki, T., Morohashi, M., Matsushima, K. & Miyawaki, T. (2000) J. Leukocyte Biol. 68, 568–574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

