An active role for tRNA in decoding beyond codon:anticodon pairing
- PMID: 15905403
- PMCID: PMC1687177
- DOI: 10.1126/science.1111408
An active role for tRNA in decoding beyond codon:anticodon pairing
Abstract
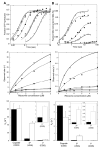
During transfer RNA (tRNA) selection, a cognate codon:anticodon interaction triggers a series of events that ultimately results in the acceptance of that tRNA into the ribosome for peptide-bond formation. High-fidelity discrimination between the cognate tRNA and near- and noncognate ones depends both on their differential dissociation rates from the ribosome and on specific acceleration of forward rate constants by cognate species. Here we show that a mutant tRNA(Trp) carrying a single substitution in its D-arm achieves elevated levels of miscoding by accelerating these forward rate constants independent of codon:anticodon pairing in the decoding center. These data provide evidence for a direct role for tRNA in signaling its own acceptance during decoding and support its fundamental role during the evolution of protein synthesis.
Figures


Comment in
-
Molecular biology. A renewed focus on transfer RNA.Science. 2005 May 20;308(5725):1123-4. doi: 10.1126/science.1113415. Science. 2005. PMID: 15905389 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

