A role for adenosine deaminase in Drosophila larval development
- PMID: 15907156
- PMCID: PMC1135298
- DOI: 10.1371/journal.pbio.0030201
A role for adenosine deaminase in Drosophila larval development
Abstract
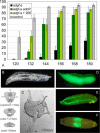
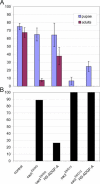
Adenosine deaminase (ADA) is an enzyme present in all organisms that catalyzes the irreversible deamination of adenosine and deoxyadenosine to inosine and deoxyinosine. Both adenosine and deoxyadenosine are biologically active purines that can have a deep impact on cellular physiology; notably, ADA deficiency in humans causes severe combined immunodeficiency. We have established a Drosophila model to study the effects of altered adenosine levels in vivo by genetic elimination of adenosine deaminase-related growth factor-A (ADGF-A), which has ADA activity and is expressed in the gut and hematopoietic organ. Here we show that the hemocytes (blood cells) are the main regulator of adenosine in the Drosophila larva, as was speculated previously for mammals. The elevated level of adenosine in the hemolymph due to lack of ADGF-A leads to apparently inconsistent phenotypic effects: precocious metamorphic changes including differentiation of macrophage-like cells and fat body disintegration on one hand, and delay of development with block of pupariation on the other. The block of pupariation appears to involve signaling through the adenosine receptor (AdoR), but fat body disintegration, which is promoted by action of the hemocytes, seems to be independent of the AdoR. The existence of such an independent mechanism has also been suggested in mammals.
Figures
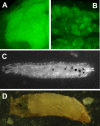
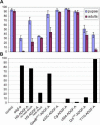
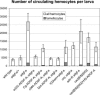

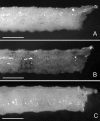

References
-
- Aldrich MB, Blackburn MR, Kellems RE. The importance of adenosine deaminase for lymphocyte development and function. Biochem Biophys Res Commun. 2000;272:311–315. - PubMed
-
- Buckley RH, Schiff RI, Schiff SE, Markert ML, Williams LW. Human severe combined immunodeficiency: Genetic, phenotypic, and functional diversity in one hundred eight infants. J Pediatr. 1997;130:378–387. - PubMed
-
- Richard E, Alam SM, Arredondo-Vega FX, Patel DD, Hershfield MS. Clustered charged amino acids of human adenosine deaminase comprise a functional epitope for binding the adenosine deaminase complexing protein CD26/dipeptidyl peptidase IV. J Biol Chem. 2002;277:19720–19726. - PubMed
-
- Latini S, Pedata F. Adenosine in the central nervous system: Release mechanisms and extracellular concentrations. J Neurochem. 2001;79:463–484. - PubMed
-
- Hershfield MS, Mitchell BS. Immunodeficiency diseases caused by adenosine deaminase deficiency and purine nucleoside phosphorylase deficiency. In: Vogelstein B, editor. The metabolic and molecular bases of inherited disease. New York: McGraw-Hill; 2001. pp. 2585–2611.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

