Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability
- PMID: 15907206
- PMCID: PMC1156890
- DOI: 10.1186/1471-2164-6-75
Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability
Abstract
Background: Microarray technology has become highly valuable for identifying complex global changes in gene expression patterns. The effective correlation of observed changes in gene expression with shared transcription regulatory elements remains difficult to demonstrate convincingly. One reason for this difficulty may result from the intricate convergence of both transcriptional and mRNA turnover events which, together, directly influence steady-state mRNA levels.
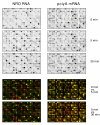
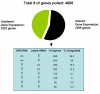
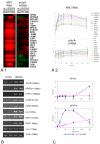
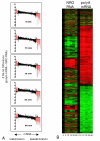
Results: In order to investigate the relative contribution of gene transcription and changes in mRNA stability regulation to standard analyses of gene expression, we used two distinct microarray methods which individually measure nuclear gene transcription and changes in polyA mRNA gene expression. Gene expression profiles were obtained from both polyA mRNA (whole-cell) and nuclear run-on (newly transcribed) RNA across a time course of one hour following the activation of human Jurkat T cells with PMA plus ionomycin. Comparative analysis revealed that regulation of mRNA stability may account for as much as 50% of all measurements of changes in polyA mRNA in this system, as inferred by the absence of any corresponding regulation of nuclear gene transcription activity for these groups of genes. Genes which displayed dramatic elevations in both mRNA and nuclear run-on RNA were shown to be inhibited by Actinomycin D (ActD) pre-treatment of cells while large numbers of genes regulated only through altered mRNA turnover (both up and down) were ActD-resistant. Consistent patterns across the time course were observed for both transcribed and stability-regulated genes.
Conclusion: We propose that regulation of mRNA stability contributes significantly to the observed changes in gene expression in response to external stimuli, as measured by high throughput systems.
Figures




References
-
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP- deficient transcription machineries. Plant J. 2002;31:171–188. doi: 10.1046/j.1365-313X.2002.01349.x. - DOI - PubMed
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
-
- Eberwine J. Amplification of mRNA populations using aRNA generated from immobilized oligo(dT)-T7 primed cDNA. Biotechniques. 1996;20:584–591. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

