Identification, distribution, and expression of novel genes in 10 clinical isolates of nontypeable Haemophilus influenzae
- PMID: 15908377
- PMCID: PMC1111819
- DOI: 10.1128/IAI.73.6.3479-3491.2005
Identification, distribution, and expression of novel genes in 10 clinical isolates of nontypeable Haemophilus influenzae
Abstract
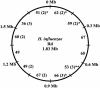
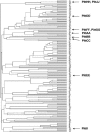
We hypothesize that Haemophilus influenzae, as a species, possesses a much greater number of genes than that found in any single H. influenzae genome. This supragenome is distributed throughout naturally occurring infectious populations, and new strains arise through autocompetence and autotransformation systems. The effect is that H. influenzae populations can readily adapt to environmental stressors. The supragenome hypothesis predicts that significant differences exist between and among the genomes of individual infectious strains of nontypeable H. influenzae (NTHi). To test this prediction, we obtained 10 low-passage NTHi clinical isolates from the middle ear effusions of patients with chronic otitis media. DNA sequencing was performed with 771 clones chosen at random from a pooled genomic library. Homology searching demonstrated that approximately 10% of these clones were novel compared to the H. influenzae Rd KW20 genome, and most of them did not match any DNA sequence in GenBank. Amino acid homology searches using hypothetical translations of the open reading frames revealed homologies to a variety of proteins, including bacterial virulence factors not previously identified in the NTHi isolates. The distribution and expression of 53 of these genes among the 10 strains were determined by PCR- and reverse transcription PCR-based analyses. These unique genes were nonuniformly distributed among the 10 isolates, and transcription of these genes in planktonic cultures was detected in 50% (177 of 352) of the occurrences. All of the novel sequences were transcribed in one or more of the NTHi isolates. Seventeen percent (9 of 53) of the novel genes were identified in all 10 NTHi strains, with each of the remaining 44 being present in only a subset of the strains. These genic distribution analyses were more effective as a strain discrimination tool than either multilocus sequence typing or 23S ribosomal gene typing methods.
Figures
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

