Target search of N sliding proteins on a DNA
- PMID: 15908574
- PMCID: PMC1366639
- DOI: 10.1529/biophysj.104.057612
Target search of N sliding proteins on a DNA
Abstract
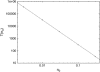
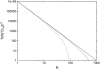
At low to moderate ambient salt concentrations, DNA-binding proteins bind relatively tightly to DNA, and only very rarely detach. Intersegmental transfer due to DNA-looping can be excluded by applying an external pulling force to the DNA molecule. Under such conditions, we explore the targeting dynamics of N proteins sliding diffusively along DNA in search of their specific target sequence. At lower densities of binding proteins, we find a reduction of the characteristic search time proportional to N(-2), with corrections at higher concentrations. Rates for detachment and attachment of binding proteins are incorporated in the model. Our findings are in agreement with recent single molecule studies in the presence of bacteriophage T4 gene 32 protein for which the unbinding rate is much lower than the specific binding rate.
Figures




Similar articles
-
First passage time of N excluded-volume particles on a line.Phys Rev E Stat Nonlin Soft Matter Phys. 2005 Oct;72(4 Pt 1):041102. doi: 10.1103/PhysRevE.72.041102. Epub 2005 Oct 17. Phys Rev E Stat Nonlin Soft Matter Phys. 2005. PMID: 16383357
-
DNA-protein interactions under random jump conditions.Phys Rev E Stat Nonlin Soft Matter Phys. 2004 Jan;69(1 Pt 1):011911. doi: 10.1103/PhysRevE.69.011911. Epub 2004 Jan 29. Phys Rev E Stat Nonlin Soft Matter Phys. 2004. PMID: 14995651
-
Base-sequence-dependent sliding of proteins on DNA.Phys Rev E Stat Nonlin Soft Matter Phys. 2004 Oct;70(4 Pt 1):041901. doi: 10.1103/PhysRevE.70.041901. Epub 2004 Oct 6. Phys Rev E Stat Nonlin Soft Matter Phys. 2004. PMID: 15600429
-
Collective dynamics of interacting molecular motors.Phys Rev Lett. 2006 Jul 21;97(3):038101. doi: 10.1103/PhysRevLett.97.038101. Epub 2006 Jul 19. Phys Rev Lett. 2006. PMID: 16907545 Review.
-
Macromolecular recognition.Curr Opin Struct Biol. 2005 Apr;15(2):171-5. doi: 10.1016/j.sbi.2005.01.018. Curr Opin Struct Biol. 2005. PMID: 15837175 Review.
Cited by
-
How DNA coiling enhances target localization by proteins.Proc Natl Acad Sci U S A. 2008 Oct 14;105(41):15738-42. doi: 10.1073/pnas.0804248105. Epub 2008 Oct 6. Proc Natl Acad Sci U S A. 2008. PMID: 18838672 Free PMC article.
-
Bulk-mediated surface transport in the presence of bias.J Chem Phys. 2017 Jul 7;147(1):014103. doi: 10.1063/1.4991730. J Chem Phys. 2017. PMID: 28688439 Free PMC article.
-
Biophysical characterization of DNA binding from single molecule force measurements.Phys Life Rev. 2010 Sep;7(3):299-341. doi: 10.1016/j.plrev.2010.06.001. Epub 2010 Jun 4. Phys Life Rev. 2010. PMID: 20576476 Free PMC article. Review.
-
In vivo facilitated diffusion model.PLoS One. 2013;8(1):e53956. doi: 10.1371/journal.pone.0053956. Epub 2013 Jan 18. PLoS One. 2013. PMID: 23349772 Free PMC article.
-
Physical constraints determine the logic of bacterial promoter architectures.Nucleic Acids Res. 2014 Apr;42(7):4196-207. doi: 10.1093/nar/gku078. Epub 2014 Jan 29. Nucleic Acids Res. 2014. PMID: 24476912 Free PMC article.
References
-
- Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell. 108:439–451. - PubMed
-
- Ptashne, M., and A. Gunn. 2002. Genes and Signals. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Kornberg, A., and T. A. Baker. 1992. DNA Replication. W. H. Freeman and Company, New York, NY.
-
- Bakk, A., and R. Metzler. 2004a. In vivo non-specific binding of λ-CI and Cro repressors is significant. FEBS Lett. 563:66–68. - PubMed
-
- Bakk, A., and R. Metzler. 2004b. Nonspecific binding of the OR repressors CI and Cro of bacteriophage λ. J. Theor. Biol. 231:525. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

