Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis
- PMID: 15908596
- PMCID: PMC1150429
- DOI: 10.1104/pp.105.061309
Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis
Abstract
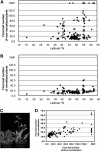
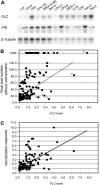
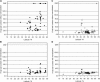
Arabidopsis (Arabidopsis thaliana) accessions provide an excellent resource to dissect the molecular basis of adaptation. We have selected 192 Arabidopsis accessions collected to represent worldwide and local variation and analyzed two adaptively important traits, flowering time and vernalization response. There was huge variation in the flowering habit of the different accessions, with no simple relationship to latitude of collection site and considerable diversity occurring within local regions. We explored the contribution to this variation from the two genes FRIGIDA (FRI) and FLOWERING LOCUS C (FLC), previously shown to be important determinants in natural variation of flowering time. A correlation of FLC expression with flowering time and vernalization was observed, but it was not as strong as anticipated due to many late-flowering/vernalization-requiring accessions being associated with low FLC expression and early-flowering accessions with high FLC expression. Sequence analysis of FRI revealed which accessions were likely to carry functional alleles, and, from comparison of flowering time with allelic type, we estimate that approximately 70% of flowering time variation can be accounted for by allelic variation of FRI. The maintenance and propagation of 20 independent nonfunctional FRI haplotypes suggest that the loss-of-function mutations can confer a strong selective advantage. Accessions with a common FRI haplotype were, in some cases, associated with very different FLC levels and wide variation in flowering time, suggesting additional variation at FLC itself or other genes regulating FLC. These data reveal how useful these Arabidopsis accessions will be in dissecting the complex molecular variation that has led to the adaptive phenotypic variation in flowering time.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

