Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis
- PMID: 15908598
- PMCID: PMC1150387
- DOI: 10.1104/pp.105.060319
Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis
Abstract
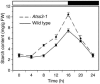
The role of starch synthase (SS) III (SSIII) in the synthesis of transient starch in Arabidopsis (Arabidopsis thaliana) was investigated by characterizing the effects of two insertion mutations at the AtSS3 gene locus. Both mutations, termed Atss3-1 and Atss3-2, condition complete loss of SSIII activity and prevent normal gene expression at both the mRNA and protein levels. The mutations cause a starch excess phenotype in leaves during the light period of the growth cycle due to an apparent increase in the rate of starch synthesis. In addition, both mutations alter the physical structure of leaf starch. Significant increases were noted in the mutants in the frequency of linear chains in amylopectin with a degree of polymerization greater than approximately 60, and relatively small changes were observed in chains of degree of polymerization 4 to 50. Furthermore, starch in the Atss3-1 and Atss3-2 mutants has a higher phosphate content, approximately two times that of wild-type leaf starch. Total SS activity is increased in both Atss3 mutants and a specific SS activity appears to be up-regulated. The data indicate that, in addition to its expected direct role in starch assembly, SSIII also has a negative regulatory function in the biosynthesis of transient starch in Arabidopsis.
Figures






Similar articles
-
Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis.BMC Plant Biol. 2008 Sep 23;8:96. doi: 10.1186/1471-2229-8-96. BMC Plant Biol. 2008. PMID: 18811962 Free PMC article.
-
Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves.Plant J. 2009 May;58(3):485-98. doi: 10.1111/j.1365-313X.2009.03793.x. Epub 2008 Jan 18. Plant J. 2009. PMID: 19154206
-
Functional and structural characterization of the catalytic domain of the starch synthase III from Arabidopsis thaliana.Proteins. 2008 Jan 1;70(1):31-40. doi: 10.1002/prot.21469. Proteins. 2008. PMID: 17623838
-
Microbial volatile-induced accumulation of exceptionally high levels of starch in Arabidopsis leaves is a process involving NTRC and starch synthase classes III and IV.Mol Plant Microbe Interact. 2011 Oct;24(10):1165-78. doi: 10.1094/MPMI-05-11-0112. Mol Plant Microbe Interact. 2011. PMID: 21649509
-
Analysis of the functional interaction of Arabidopsis starch synthase and branching enzyme isoforms reveals that the cooperative action of SSI and BEs results in glucans with polymodal chain length distribution similar to amylopectin.PLoS One. 2014 Jul 11;9(7):e102364. doi: 10.1371/journal.pone.0102364. eCollection 2014. PLoS One. 2014. PMID: 25014622 Free PMC article.
Cited by
-
Tuning heterologous glucan biosynthesis in yeast to understand and exploit plant starch diversity.BMC Biol. 2022 Sep 24;20(1):207. doi: 10.1186/s12915-022-01408-x. BMC Biol. 2022. PMID: 36153520 Free PMC article.
-
Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes.Plant Physiol. 2008 Apr;146(4):1878-91. doi: 10.1104/pp.108.116244. Epub 2008 Feb 8. Plant Physiol. 2008. PMID: 18263778 Free PMC article.
-
The barley amo1 locus is tightly linked to the starch synthase IIIa gene and negatively regulates expression of granule-bound starch synthetic genes.J Exp Bot. 2011 Oct;62(14):5217-31. doi: 10.1093/jxb/err239. Epub 2011 Aug 3. J Exp Bot. 2011. PMID: 21813797 Free PMC article.
-
Transcriptomics and starch biosynthesis analysis in leaves and developing seeds of mung bean provide a basis for genetic engineering of starch composition and seed quality.Front Plant Sci. 2024 May 1;15:1332150. doi: 10.3389/fpls.2024.1332150. eCollection 2024. Front Plant Sci. 2024. PMID: 38751837 Free PMC article.
-
Distinct Functions of STARCH SYNTHASE 4 Domains in Starch Granule Formation.Plant Physiol. 2018 Jan;176(1):566-581. doi: 10.1104/pp.17.01008. Epub 2017 Nov 13. Plant Physiol. 2018. PMID: 29133376 Free PMC article.
References
-
- Abel GJW, Springer F, Willmitzer L, Kossmann J (1996) Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato (Solanum tuberosum L.). Plant J 10: 981–991 - PubMed
-
- Blennow A, Engelsen SB, Munck L, Moller BL (2000) Starch molecular structure and phosphorylation investigated by a combined chromatographic and chemometric approach. Carbohydr Polym 41: 163–174
-
- Blennow A, Nielsen TH, Baunsgaard L, Mikkelsen R, Engelsen SB (2002) Starch phosphorylation: a new front line in starch research. Trends Plant Sci 7: 445–450 - PubMed
-
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

