Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM
- PMID: 15908964
- PMCID: PMC1925259
- DOI: 10.1038/nsmb938
Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM
Abstract
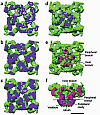
RyR1 is an intracellular calcium channel with a central role in muscle contraction. We obtained a three-dimensional reconstruction of the RyR1 in the closed state at a nominal resolution of approximately 10 A using cryo-EM. The cytoplasmic assembly consists of a series of interconnected tubular structures that merge into four columns that extend into the transmembrane assembly. The transmembrane assembly, which has at least six transmembrane alpha-helices per monomer, has four tilted rods that can be fitted with the inner helices of a closed K(+) channel atomic structure. The rods splay out at the lumenal side and converge into a dense ring at the cytoplasmic side. Another set of four rods emerges from this ring and shapes the inner part of the four columns. The resulting constricted axial structure provides direct continuity between cytoplasmic and transmembrane assemblies, and a possible mechanism for control of channel gating through conformational changes in the cytoplasmic assembly.
Figures
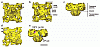
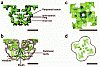

References
-
- Fill M, Copello JA. Ryanodine receptor calcium release channels. Physiol. Rev. 2002;82:893–922. - PubMed
-
- Dulhunty AF, Pouliquin P. What we don’t know about the structure of ryanodine receptor calcium release channels. Clin. Exp. Pharmacol. Physiol. 2003;30:713–723. - PubMed
-
- Wagenknecht T, Samsoó M. Three‐dimensional reconstruction of ryanodine receptors. Front. Biosci. 2002;7:d1464–d1474. - PubMed
-
- Wolf M, Eberhart A, Glossmann H, Striessnig J, Grigorieff N. Visualization of the domain structure of an L‐type Ca2+ channel using electron cryo‐microscopy. J. Mol. Biol. 2003;332:171–182. - PubMed
-
- Paolini C, Protasi F, Franzini‐Armstrong C. The relative position of RyR feet and DHPR tetrads in skeletal muscle. J. Mol. Biol. 2004;342:145–153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

