Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease
- PMID: 15911761
- PMCID: PMC1142368
- DOI: 10.1073/pnas.0501078102
Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease
Abstract
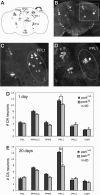
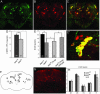
Loss-of-function mutations of the parkin gene are a major cause of early-onset parkinsonism. To explore the mechanism by which loss of parkin function results in neurodegeneration, we are using a genetic approach in Drosophila. Here, we show that Drosophila parkin mutants display degeneration of a subset of dopaminergic (DA) neurons in the brain. The neurodegenerative phenotype of parkin mutants is enhanced by loss-of-function mutations of the glutathione S-transferase S1 (GstS1) gene, which were identified in an unbiased genetic screen for genes that modify parkin phenotypes. Furthermore, overexpression of GstS1 in DA neurons suppresses neurodegeneration in parkin mutants. Given the previous evidence for altered glutathione metabolism and oxidative stress in sporadic Parkinson's disease (PD), these data suggest that the mechanism of DA neuron loss in Drosophila parkin mutants is similar to the mechanisms underlying sporadic PD. Moreover, these findings identify a potential therapeutic approach in treating PD.
Figures


Similar articles
-
Glutathione S-transferase omega suppresses the defective phenotypes caused by PINK1 loss-of-function in Drosophila.Biochem Biophys Res Commun. 2013 Aug 9;437(4):615-9. doi: 10.1016/j.bbrc.2013.07.011. Epub 2013 Jul 15. Biochem Biophys Res Commun. 2013. PMID: 23867819
-
Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin.Nature. 2006 Jun 29;441(7097):1162-6. doi: 10.1038/nature04779. Epub 2006 May 3. Nature. 2006. PMID: 16672981
-
Natural variation in age-related dopamine neuron degeneration is glutathione dependent and linked to life span.Proc Natl Acad Sci U S A. 2024 Oct 15;121(42):e2403450121. doi: 10.1073/pnas.2403450121. Epub 2024 Oct 10. Proc Natl Acad Sci U S A. 2024. PMID: 39388265 Free PMC article.
-
[Etiology and pathogenesis of Parkinson's disease: from mitochondrial dysfunctions to familial Parkinson's disease].Rinsho Shinkeigaku. 2004 Apr-May;44(4-5):241-62. Rinsho Shinkeigaku. 2004. PMID: 15287506 Review. Japanese.
-
The synaptic function of parkin.Brain. 2017 Sep 1;140(9):2265-2272. doi: 10.1093/brain/awx006. Brain. 2017. PMID: 28335015 Review.
Cited by
-
Sequestration and autophagy of mitochondria do not cut proteins across the board.Proc Natl Acad Sci U S A. 2013 Apr 16;110(16):6252-3. doi: 10.1073/pnas.1303921110. Epub 2013 Apr 8. Proc Natl Acad Sci U S A. 2013. PMID: 23569251 Free PMC article. No abstract available.
-
Liver-specific Prkn knockout mice are more susceptible to diet-induced hepatic steatosis and insulin resistance.Mol Metab. 2020 Nov;41:101051. doi: 10.1016/j.molmet.2020.101051. Epub 2020 Jul 10. Mol Metab. 2020. PMID: 32653576 Free PMC article.
-
Parkinson-related parkin reduces α-Synuclein phosphorylation in a gene transfer model.Mol Neurodegener. 2010 Nov 4;5:47. doi: 10.1186/1750-1326-5-47. Mol Neurodegener. 2010. PMID: 21050448 Free PMC article.
-
Glial dysfunction in parkin null mice: effects of aging.J Neurosci. 2008 Jan 16;28(3):598-611. doi: 10.1523/JNEUROSCI.4609-07.2008. J Neurosci. 2008. PMID: 18199761 Free PMC article.
-
Vulnerable Parkin Loss-of-Function Drosophila Dopaminergic Neurons Have Advanced Mitochondrial Aging, Mitochondrial Network Loss and Transiently Reduced Autophagosome Recruitment.Front Cell Neurosci. 2018 Feb 15;12:39. doi: 10.3389/fncel.2018.00039. eCollection 2018. Front Cell Neurosci. 2018. PMID: 29497364 Free PMC article.
References
-
- Dawson, T. M. & Dawson, V. L. (2003) Science 302, 819–822. - PubMed
-
- West, A. B. & Maidment, N. T. (2004) Hum. Genet. 114, 327–336. - PubMed
-
- Chung, K. K., Thomas, B., Li, X., Pletnikova, O., Troncoso, J. C., Marsh, L., Dawson, V. L. & Dawson, T. M. (2004) Science 304, 1328–1331. - PubMed
-
- Pawlyk, A. C., Giasson, B. I., Sampathu, D. M., Perez, F. A., Lim, K. L., Dawson, V. L., Dawson, T. M., Palmiter, R. D., Trojanowski, J. Q. & Lee, V. M. (2003) J. Biol. Chem. 278, 48120–48128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

