Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation
- PMID: 15914556
- PMCID: PMC1895212
- DOI: 10.1182/blood-2005-03-0860
Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation
Abstract
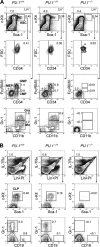
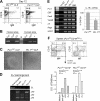
The PU.1 transcription factor is a key regulator of hematopoietic development, but its role at each hematopoietic stage remains unclear. In particular, the expression of PU.1 in hematopoietic stem cells (HSCs) could simply represent "priming" of genes related to downstream myelolymphoid lineages. By using a conditional PU.1 knock-out model, we here show that HSCs express PU.1, and its constitutive expression is necessary for maintenance of the HSC pool in the bone marrow. Bone marrow HSCs disrupted with PU.1 in situ could not maintain hematopoiesis and were outcompeted by normal HSCs. PU.1-deficient HSCs also failed to generate the earliest myeloid and lymphoid progenitors. PU.1 disruption in granulocyte/monocyte-committed progenitors blocked their maturation but not proliferation, resulting in myeloblast colony formation. PU.1 disruption in common lymphoid progenitors, however, did not prevent their B-cell maturation. In vivo disruption of PU.1 in mature B cells by the CD19-Cre locus did not affect B-cell maturation, and PU.1-deficient mature B cells displayed normal proliferation in response to mitogenic signals including the cross-linking of surface immunoglobulin M (IgM). Thus, PU.1 plays indispensable and distinct roles in hematopoietic development through supporting HSC self-renewal as well as commitment and maturation of myeloid and lymphoid lineages.
Figures

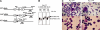



References
-
- Shivdasani RA, Orkin SH. The transcriptional control of hematopoiesis. Blood. 1996;87: 4025-4039. - PubMed
-
- Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90: 489-519. - PubMed
-
- Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61: 113-124. - PubMed
-
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404: 193-197. - PubMed
-
- Miyamoto T, Iwasaki H, Reizis B, et al. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell. 2002;3: 137-147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

