Prevalence of quadruplexes in the human genome
- PMID: 15914667
- PMCID: PMC1140081
- DOI: 10.1093/nar/gki609
Prevalence of quadruplexes in the human genome
Abstract
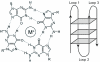
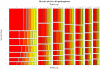
Guanine-rich DNA sequences of a particular form have the ability to fold into four-stranded structures called G-quadruplexes. In this paper, we present a working rule to predict which primary sequences can form this structure, and describe a search algorithm to identify such sequences in genomic DNA. We count the number of quadruplexes found in the human genome and compare that with the figure predicted by modelling DNA as a Bernoulli stream or as a Markov chain, using windows of various sizes. We demonstrate that the distribution of loop lengths is significantly different from what would be expected in a random case, providing an indication of the number of potentially relevant quadruplex-forming sequences. In particular, we show that there is a significant repression of quadruplexes in the coding strand of exonic regions, which suggests that quadruplex-forming patterns are disfavoured in sequences that will form RNA.
Figures

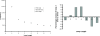
References
-
- Guschlbauer W., Chantot J.F., Theile D. Four-stranded nucleic structures 25 years later: from guanosine gels to telomere DNA. J. Biomol. Struct. Dyn. 1990;8:491–511. - PubMed
-
- Blackburn E.H. Telomeres and their synthesis. Science. 1990;249:489–490. - PubMed
-
- Blackburn E.H. Structure and function of telomeres. Nature. 1991;350:569–573. - PubMed
-
- Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

