Norfloxacin-induced DNA gyrase cleavage complexes block Escherichia coli replication forks, causing double-stranded breaks in vivo
- PMID: 15916595
- PMCID: PMC1201555
- DOI: 10.1111/j.1365-2958.2005.04638.x
Norfloxacin-induced DNA gyrase cleavage complexes block Escherichia coli replication forks, causing double-stranded breaks in vivo
Abstract
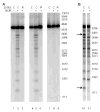
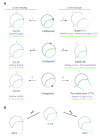
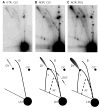
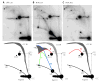
Antibacterial quinolones inhibit type II DNA topoisomerases by stabilizing covalent topoisomerase-DNA cleavage complexes, which are apparently transformed into double-stranded breaks by cellular processes such as replication. We used plasmid pBR322 and two-dimensional agarose gel electrophoresis to examine the collision of replication forks with quinolone-induced gyrase-DNA cleavage complexes in Escherichia coli. Restriction endonuclease-digested DNA exhibited a bubble arc with discrete spots, indicating that replication forks had been stalled. The most prominent spot depended upon the strong gyrase binding site of pBR322, providing direct evidence that quinolone-induced cleavage complexes block bacterial replication forks in vivo. We differentiated between stalled forks that do or do not contain bound cleavage complex by extracting DNA under different conditions. Resealing conditions allow gyrase to efficiently reseal the transient breaks within cleavage complexes, while cleavage conditions cause the latent breaks to be revealed. These experiments showed that some stalled forks did not contain a cleavage complex, implying that gyrase had dissociated in vivo and yet the fork had not restarted at the time of DNA isolation. Additionally, some branched plasmid DNA isolated under resealing conditions nonetheless contained broken DNA ends. We discuss a model for the creation of double-stranded breaks by an indirect mechanism after quinolone treatment.
Figures


Similar articles
-
DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage.J Mol Biol. 1996 May 17;258(4):627-37. doi: 10.1006/jmbi.1996.0274. J Mol Biol. 1996. PMID: 8636997
-
An antitumor drug-induced topoisomerase cleavage complex blocks a bacteriophage T4 replication fork in vivo.Mol Cell Biol. 2000 Jan;20(2):594-603. doi: 10.1128/MCB.20.2.594-603.2000. Mol Cell Biol. 2000. PMID: 10611238 Free PMC article.
-
Isolation and quantitation of topoisomerase complexes accumulated on Escherichia coli chromosomal DNA.Antimicrob Agents Chemother. 2012 Nov;56(11):5458-64. doi: 10.1128/AAC.01182-12. Epub 2012 Aug 6. Antimicrob Agents Chemother. 2012. PMID: 22869559 Free PMC article.
-
Mode of action of the new quinolones: new data.Eur J Clin Microbiol Infect Dis. 1991 Apr;10(4):223-31. doi: 10.1007/BF01966994. Eur J Clin Microbiol Infect Dis. 1991. PMID: 1650698 Review.
-
Molecular interactions of the CcdB poison with its bacterial target, the DNA gyrase.Int J Med Microbiol. 2002 Feb;291(6-7):537-44. doi: 10.1078/1438-4221-00164. Int J Med Microbiol. 2002. PMID: 11890555 Review.
Cited by
-
Effect of anaerobic growth on quinolone lethality with Escherichia coli.Antimicrob Agents Chemother. 2007 Jan;51(1):28-34. doi: 10.1128/AAC.00739-06. Epub 2006 Oct 16. Antimicrob Agents Chemother. 2007. PMID: 17043118 Free PMC article.
-
RecA can stimulate the relaxation activity of topoisomerase I: Molecular basis of topoisomerase-mediated genome-wide transcriptional responses in Escherichia coli.Nucleic Acids Res. 2007;35(1):79-86. doi: 10.1093/nar/gkl981. Epub 2006 Dec 6. Nucleic Acids Res. 2007. PMID: 17151069 Free PMC article.
-
Growth-dependent heterogeneity in the DNA damage response in Escherichia coli.Mol Syst Biol. 2022 May;18(5):e10441. doi: 10.15252/msb.202110441. Mol Syst Biol. 2022. PMID: 35620827 Free PMC article.
-
TisB protein is the single molecular determinant underlying multiple downstream effects of ofloxacin in Escherichia coli.Sci Adv. 2024 Mar 29;10(13):eadk1577. doi: 10.1126/sciadv.adk1577. Epub 2024 Mar 27. Sci Adv. 2024. PMID: 38536908 Free PMC article.
-
Fluoroquinolone-gyrase-DNA complexes: two modes of drug binding.J Biol Chem. 2014 May 2;289(18):12300-12. doi: 10.1074/jbc.M113.529164. Epub 2014 Feb 4. J Biol Chem. 2014. PMID: 24497635 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

