SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli
- PMID: 15916962
- PMCID: PMC4428309
- DOI: 10.1016/j.molcel.2005.04.012
SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli
Abstract
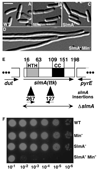
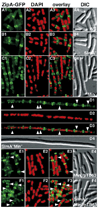
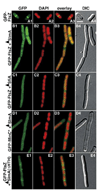
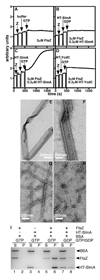
Cell division in Escherichia coli begins with assembly of the tubulin-like FtsZ protein into a ring structure just underneath the cell membrane. Spatial control over Z ring assembly is achieved by two partially redundant negative regulatory systems, the Min system and nucleoid occlusion (NO), which cooperate to position the division site at midcell. In contrast to the well-studied Min system, almost nothing is known about how Z ring assembly is blocked in the vicinity of nucleoids to effect NO. Reasoning that Min function might become essential in cells impaired for NO, we screened for mutations synthetically lethal with a defective Min system (slm mutants). By using this approach, we identified SlmA (Ttk) as the first NO factor in E. coli. Our combined genetic, cytological, and biochemical results suggest that SlmA is a DNA-associated division inhibitor that is directly involved in preventing Z ring assembly on portions of the membrane surrounding the nucleoid.
Figures


References
-
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 2005;55:1631–1645. - PubMed
-
- Aussel L, Barre F-X, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

