Natural and synthetic tetracycline-inducible promoters for use in the antibiotic-producing bacteria Streptomyces
- PMID: 15917435
- PMCID: PMC1140374
- DOI: 10.1093/nar/gni086
Natural and synthetic tetracycline-inducible promoters for use in the antibiotic-producing bacteria Streptomyces
Abstract
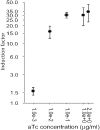
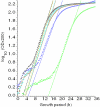
Bacteria in the genus Streptomyces are major producers of antibiotics and other pharmacologically active compounds. Genetic and physiological manipulations of these bacteria are important for new drug discovery and production development. An essential part of any 'genetic toolkit' is the availability of regulatable promoters. We have adapted the tetracycline (Tc) repressor/operator (TetR/tetO) regulatable system from transposon Tn10 for use in Streptomyces. The synthetic Tc controllable promoter (tcp), tcp830, was active in a wide range of Streptomyces species, and varying levels of induction were observed after the addition of 1-100 ng/ml of anhydrotetracycline (aTc). Streptomyces coelicolor contained an innate Tc-controllable promoter regulated by a TetR homologue (SCO0253). Both natural and synthetic promoters were active and inducible throughout growth. Using the luxAB genes expressing luciferase as a reporter system, we showed that induction factors of up to 270 could be obtained for tcp830. The effect of inducers on the growth of S.coelicolor was determined; addition of aTc at concentrations where induction is optimal, i.e. 0.1-1 microg/ml, ranged from no effect on growth rate to a small increase in the lag period compared with cultures with no inducer.
Figures


Similar articles
-
Tetracycline-inducible gene expression in mycobacteria within an animal host using modified Streptomyces tcp830 regulatory elements.Arch Microbiol. 2006 Dec;186(6):459-64. doi: 10.1007/s00203-006-0160-2. Epub 2006 Aug 30. Arch Microbiol. 2006. PMID: 16944099
-
Tight control of transcription in Toxoplasma gondii using an alternative tet repressor.Int J Parasitol. 2006 Apr;36(4):443-52. doi: 10.1016/j.ijpara.2006.01.005. Epub 2006 Feb 13. Int J Parasitol. 2006. PMID: 16516216
-
Application of redD, the transcriptional activator gene of the undecylprodigiosin biosynthetic pathway, as a reporter for transcriptional activity in Streptomyces coelicolor A3(2) and Streptomyces lividans.J Mol Microbiol Biotechnol. 2000 Oct;2(4):551-6. J Mol Microbiol Biotechnol. 2000. PMID: 11075931
-
Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor.Mol Microbiol. 2006 Feb;59(3):731-42. doi: 10.1111/j.1365-2958.2005.05018.x. Mol Microbiol. 2006. PMID: 16420347 Review.
-
The application of Tet repressor in prokaryotic gene regulation and expression.Microb Biotechnol. 2008 Jan;1(1):2-16. doi: 10.1111/j.1751-7915.2007.00001.x. Microb Biotechnol. 2008. PMID: 21261817 Free PMC article. Review.
Cited by
-
Current Approaches for Genetic Manipulation of Streptomyces spp.-Key Bacteria for Biotechnology and Environment.BioTech (Basel). 2025 Jan 2;14(1):3. doi: 10.3390/biotech14010003. BioTech (Basel). 2025. PMID: 39846552 Free PMC article. Review.
-
Novel switchable ECF sigma factor transcription system for improving thaxtomin A production in Streptomyces.Synth Syst Biotechnol. 2022 Jun 6;7(3):972-981. doi: 10.1016/j.synbio.2022.05.010. eCollection 2022 Sep. Synth Syst Biotechnol. 2022. PMID: 35756964 Free PMC article.
-
sRNA scr5239 Involved in Feedback Loop Regulation of Streptomyces coelicolor Central Metabolism.Front Microbiol. 2020 Jan 23;10:3121. doi: 10.3389/fmicb.2019.03121. eCollection 2019. Front Microbiol. 2020. PMID: 32117084 Free PMC article.
-
A tunable and reversible thermo-inducible bio-switch for streptomycetes.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1236. doi: 10.1093/nar/gkae1236. Nucleic Acids Res. 2025. PMID: 39704119 Free PMC article.
-
Activation of a Cryptic Manumycin-Type Biosynthetic Gene Cluster of Saccharothrix espanaensis DSM44229 by Series of Genetic Manipulations.Microorganisms. 2021 Mar 8;9(3):559. doi: 10.3390/microorganisms9030559. Microorganisms. 2021. PMID: 33800500 Free PMC article.
References
-
- Ali N., Herron P.R., Evans M.C., Dyson P.J. Osmotic regulation of the Streptomyces lividans thiostrepton-inducible promoter, ptipA. Microbiology. 2002;148:381–390. - PubMed
-
- Weaden J., Dyson P. Transposon mutagenesis with IS6100 in the avermectin-producer Streptomyces avermitilis. Microbiology. 1998;144:1963–1970. - PubMed
-
- Hindle Z., Smith C.P. Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol. Microbiol. 1994;12:737–745. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

