Conservation of functional domains and limited heterogeneity of HIV-1 reverse transcriptase gene following vertical transmission
- PMID: 15918905
- PMCID: PMC1166575
- DOI: 10.1186/1742-4690-2-36
Conservation of functional domains and limited heterogeneity of HIV-1 reverse transcriptase gene following vertical transmission
Abstract
Background: The reverse transcriptase (RT) enzyme of human immunodeficiency virus type 1 (HIV-1) plays a crucial role in the life cycle of the virus by converting the single stranded RNA genome into double stranded DNA that integrates into the host chromosome. In addition, RT is also responsible for the generation of mutations throughout the viral genome, including in its own sequences and is thus responsible for the generation of quasi-species in HIV-1-infected individuals. We therefore characterized the molecular properties of RT, including the conservation of functional motifs, degree of genetic diversity, and evolutionary dynamics from five mother-infant pairs following vertical transmission.
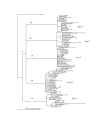
Results: The RT open reading frame was maintained with a frequency of 87.2% in five mother-infant pairs' sequences following vertical transmission. There was a low degree of viral heterogeneity and estimates of genetic diversity in mother-infant pairs' sequences. Both mothers and infants RT sequences were under positive selection pressure, as determined by the ratios of non-synonymous to synonymous substitutions. Phylogenetic analysis of 132 mother-infant RT sequences revealed distinct clusters for each mother-infant pair, suggesting that the epidemiologically linked mother-infant pairs were evolutionarily closer to each other as compared with epidemiologically unlinked mother-infant pairs. The functional domains of RT which are responsible for reverse transcription, DNA polymerization and RNase H activity were mostly conserved in the RT sequences analyzed in this study. Specifically, the active sites and domains required for primer binding, template binding, primer and template positioning and nucleotide recruitment were conserved in all mother-infant pairs' sequences.
Conclusion: The maintenance of an intact RT open reading frame, conservation of functional domains for RT activity, preservation of several amino acid motifs in epidemiologically linked mother-infant pairs, and a low degree of genetic variability following vertical transmission is consistent with an indispensable role of RT in HIV-1 replication in infected mother-infant pairs.
Figures






Similar articles
-
Characterization of HIV-1 envelope gp41 genetic diversity and functional domains following perinatal transmission.Retrovirology. 2006 Jul 4;3:42. doi: 10.1186/1742-4690-3-42. Retrovirology. 2006. PMID: 16820061 Free PMC article.
-
Molecular characterization of the HIV-1 gag nucleocapsid gene associated with vertical transmission.Retrovirology. 2006 Apr 6;3:21. doi: 10.1186/1742-4690-3-21. Retrovirology. 2006. PMID: 16600029 Free PMC article.
-
Evaluations of HIV type 1 rev gene diversity and functional domains following perinatal transmission.AIDS Res Hum Retroviruses. 2005 Dec;21(12):1035-45. doi: 10.1089/aid.2005.21.1035. AIDS Res Hum Retroviruses. 2005. PMID: 16379607
-
The vertical transmission of human immunodeficiency virus type 1: molecular and biological properties of the virus.Crit Rev Clin Lab Sci. 2005;42(1):1-34. doi: 10.1080/10408360490512520. Crit Rev Clin Lab Sci. 2005. PMID: 15697169 Review.
-
Human Immunodeficiency Virus type 1 reverse transcriptase.Biol Chem Hoppe Seyler. 1996 Feb;377(2):97-120. Biol Chem Hoppe Seyler. 1996. PMID: 8868066 Review.
Cited by
-
Characterization of HIV-1 envelope gp41 genetic diversity and functional domains following perinatal transmission.Retrovirology. 2006 Jul 4;3:42. doi: 10.1186/1742-4690-3-42. Retrovirology. 2006. PMID: 16820061 Free PMC article.
-
Features of Maternal HIV-1 Associated with Lack of Vertical Transmission.Open Virol J. 2017 Mar 23;11:8-14. doi: 10.2174/1874357901710011008. eCollection 2017. Open Virol J. 2017. PMID: 28458735 Free PMC article. Review.
-
Short communication: Nucleotide variation and positively selected sites in HIV type 1 reverse transcriptase among heterosexual transmission pairs.AIDS Res Hum Retroviruses. 2010 Aug;26(8):895-9. doi: 10.1089/aid.2010.0003. AIDS Res Hum Retroviruses. 2010. PMID: 20672974 Free PMC article.
-
Sagacious epitope selection for vaccines, and both antibody-based therapeutics and diagnostics: tips from virology and oncology.Antib Ther. 2022 Feb 21;5(1):63-72. doi: 10.1093/abt/tbac005. eCollection 2022 Jan. Antib Ther. 2022. PMID: 35372784 Free PMC article. Review.
-
Molecular characterization of the HIV-1 gag nucleocapsid gene associated with vertical transmission.Retrovirology. 2006 Apr 6;3:21. doi: 10.1186/1742-4690-3-21. Retrovirology. 2006. PMID: 16600029 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

