Comparative analysis of chromatin landscape in regulatory regions of human housekeeping and tissue specific genes
- PMID: 15918906
- PMCID: PMC1173084
- DOI: 10.1186/1471-2105-6-126
Comparative analysis of chromatin landscape in regulatory regions of human housekeeping and tissue specific genes
Abstract
Background: Global regulatory mechanisms involving chromatin assembly and remodelling in the promoter regions of genes is implicated in eukaryotic transcription control especially for genes subjected to spatial and temporal regulation. The potential to utilise global regulatory mechanisms for controlling gene expression might depend upon the architecture of the chromatin in and around the gene. In-silico analysis can yield important insights into this aspect, facilitating comparison of two or more classes of genes comprising of a large number of genes within each group.
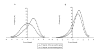
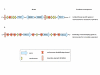
Results: In the present study, we carried out a comparative analysis of chromatin characteristics in terms of the scaffold/matrix attachment regions, nucleosome formation potential and the occurrence of repetitive sequences, in the upstream regulatory regions of housekeeping and tissue specific genes. Our data show that putative scaffold/matrix attachment regions are more abundant and nucleosome formation potential is higher in the 5' regions of tissue specific genes as compared to the housekeeping genes.
Conclusion: The differences in the chromatin features between the two groups of genes indicate the involvement of chromatin organisation in the control of gene expression. The presence of global regulatory mechanisms mediated through chromatin organisation can decrease the burden of invoking gene specific regulators for maintenance of the active/silenced state of gene expression. This could partially explain the lower number of genes estimated in the human genome.
Figures
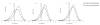
References
-
- Bode J, Benham C, Knopp A, Mielke C. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements) Crit Rev Eukaryot Gene Expr. 2000;10:73–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

