The limits of protein sequence comparison?
- PMID: 15919194
- PMCID: PMC2845305
- DOI: 10.1016/j.sbi.2005.05.005
The limits of protein sequence comparison?
Abstract
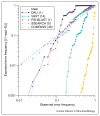
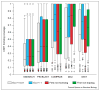
Modern sequence alignment algorithms are used routinely to identify homologous proteins, proteins that share a common ancestor. Homologous proteins always share similar structures and often have similar functions. Over the past 20 years, sequence comparison has become both more sensitive, largely because of profile-based methods, and more reliable, because of more accurate statistical estimates. As sequence and structure databases become larger, and comparison methods become more powerful, reliable statistical estimates will become even more important for distinguishing similarities that are due to homology from those that are due to analogy (convergence). The newest sequence alignment methods are more sensitive than older methods, but more accurate statistical estimates are needed for their full power to be realized.
Figures

References
-
- Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Apweiler R, Bairoch A, Wu CH. Protein sequence databases. Curr Opin Chem Biol. 2004;8:76–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

