Analysis and function of prototype foamy virus envelope N glycosylation
- PMID: 15919919
- PMCID: PMC1143653
- DOI: 10.1128/JVI.79.12.7664-7672.2005
Analysis and function of prototype foamy virus envelope N glycosylation
Abstract
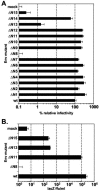
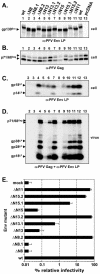
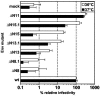
The prototype foamy virus (PFV) glycoprotein, which is essential for PFV particle release, displays a highly unusual biosynthesis, resulting in posttranslational cleavage of the precursor protein into three particle-associated subunits, i.e., leader peptide (LP), surface (SU), and transmembrane (TM). Glycosidase digestion of metabolically labeled PFV particles revealed the presence of N-linked carbohydrates on all subunits. The differential sensitivity to specific glycosidases indicated that all oligosaccharides on LP and TM are of the high-mannose or hybrid type, whereas most of those attached to SU, which contribute to about 50% of its molecular weight, are of the complex type. Individual inactivation of all 15 potential N-glycosylation sites in PFV Env demonstrated that 14 are used, i.e., 1 out of 2 in LP, 10 in SU, and 3 in TM. Analysis of the individual altered glycoproteins revealed defects in intracellular processing, support of particle release, and infectivity for three mutants, having the evolutionarily conserved glycosylation sites N8 in SU or N13 and N15 in the cysteine-rich central "sheets-and-loops" region of TM inactivated. Examination of alternative mutants with mutations affecting glycosylation or surrounding sequences at these sites indicated that inhibition of glycosylation at N8 and N13 most likely is responsible for the observed replication defects, whereas for N15 surrounding sequences seem to contribute to a temperature-sensitive phenotype. Taken together these data demonstrate that PFV Env and in particular the SU subunit are heavily N glycosylated and suggest that although most carbohydrates are dispensable individually, some evolutionarily conserved sites are important for normal Env function of FV isolates from different species.
Figures




Similar articles
-
Characterization of the prototype foamy virus envelope glycoprotein receptor-binding domain.J Virol. 2006 Aug;80(16):8158-67. doi: 10.1128/JVI.00460-06. J Virol. 2006. PMID: 16873272 Free PMC article.
-
Ubiquitination of the prototype foamy virus envelope glycoprotein leader peptide regulates subviral particle release.J Virol. 2005 Dec;79(24):15074-83. doi: 10.1128/JVI.79.24.15074-15083.2005. J Virol. 2005. PMID: 16306578 Free PMC article.
-
Prototype foamy virus envelope glycoprotein leader peptide processing is mediated by a furin-like cellular protease, but cleavage is not essential for viral infectivity.J Virol. 2004 Dec;78(24):13865-70. doi: 10.1128/JVI.78.24.13865-13870.2004. J Virol. 2004. PMID: 15564494 Free PMC article.
-
The foamy virus envelope glycoproteins.Curr Top Microbiol Immunol. 2003;277:111-29. doi: 10.1007/978-3-642-55701-9_5. Curr Top Microbiol Immunol. 2003. PMID: 12908770 Review.
-
The HTLV-I envelope glycoproteins: structure and functions.J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13 Suppl 1:S85-91. doi: 10.1097/00042560-199600001-00015. J Acquir Immune Defic Syndr Hum Retrovirol. 1996. PMID: 8797709 Review.
Cited by
-
Molecular biology of foamy viruses.Med Microbiol Immunol. 2010 Aug;199(3):197-207. doi: 10.1007/s00430-010-0158-x. Epub 2010 May 6. Med Microbiol Immunol. 2010. PMID: 20445989 Review.
-
Neutralization of zoonotic retroviruses by human antibodies: Genotype-specific epitopes within the receptor-binding domain from simian foamy virus.PLoS Pathog. 2023 Apr 24;19(4):e1011339. doi: 10.1371/journal.ppat.1011339. eCollection 2023 Apr. PLoS Pathog. 2023. PMID: 37093892 Free PMC article.
-
Efficient production of inhibitor-free foamy virus glycoprotein-containing retroviral vectors by proteoglycan-deficient packaging cells.Mol Ther Methods Clin Dev. 2022 Aug 1;26:394-412. doi: 10.1016/j.omtm.2022.07.004. eCollection 2022 Sep 8. Mol Ther Methods Clin Dev. 2022. PMID: 36034773 Free PMC article.
-
The Unique, the Known, and the Unknown of Spumaretrovirus Assembly.Viruses. 2021 Jan 13;13(1):105. doi: 10.3390/v13010105. Viruses. 2021. PMID: 33451128 Free PMC article. Review.
-
Modular nature of simian foamy virus genomes and their evolutionary history.Virus Evol. 2019 Oct 16;5(2):vez032. doi: 10.1093/ve/vez032. eCollection 2019 Jul. Virus Evol. 2019. PMID: 31636999 Free PMC article.
References
-
- Doms, R. W., R. A. Lamb, J. K. Rose, and A. Helenius. 1993. Folding and assembly of viral membrane proteins. Virology 193:545-562. - PubMed
-
- Duda, A., A. Stange, D. Lüftenegger, N. Stanke, D. Westphal, T. Pietschmann, S. W. Eastman, M. L. Linial, A. Rethwilm, and D. Lindemann. 2004. Prototype foamy virus envelope glycoprotein leader peptide processing is mediated by a furin-like cellular protease, but cleavage is not essential for viral infectivity. J. Virol. 78:13865-13870. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

