Single amino acid insertions at the junction of the sindbis virus E2 transmembrane domain and endodomain disrupt virus envelopment and alter infectivity
- PMID: 15919921
- PMCID: PMC1143637
- DOI: 10.1128/JVI.79.12.7682-7697.2005
Single amino acid insertions at the junction of the sindbis virus E2 transmembrane domain and endodomain disrupt virus envelopment and alter infectivity
Abstract
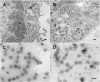
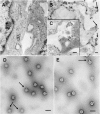
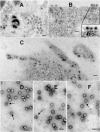
The final steps in the envelopment of Sindbis virus involve specific interactions of the E2 endodomain with the virus nucleocapsid. Deleting E2 K at position 391 (E2 DeltaK391) resulted in the disruption of virus assembly in mammalian cells but not insect cells (host range mutant). This suggested unique interactions of the E2 DeltaK391 endodomain with the different biochemical environments of the mammalian and insect cell lipid bilayers. To further investigate the role of the amino acid residues located at or around position E2 391 and constraints on the length of the endodomain on virus assembly, amino acid insertions/substitutions at the transmembrane/endodomain junction were constructed. An additional K was inserted at amino acid position 392 (KK391/392), a K-->F substitution at position 391 was constructed (F391), and an additional F was inserted at 392 (FF391/392). These changes should lengthen the endodomain in the KK391/392 insertion mutant or shorten the endodomain in the FF391/392 mutant. The mutant FF391/392 grown in BHK cells formed virus particles containing extruded material not found on wild-type virus. This characteristic was not seen in FF391/392 virus grown in insect cells. The mutant KK391/392 grown in BHK cells was defective in the final membrane fission reaction, producing multicored or conjoined virus particles. The production of these aberrant particles was ameliorated when the KK391/392 mutant was grown in insect cells. These data indicate that there is a critical minimal spanning distance from the E2 membrane proximal amino acid at position 391 and the conserved E2 Y400 residue. The observed phenotypes of these mutants also invoke an important role of the specific host membrane lipid composition on virus architecture and infectivity.
Figures

Similar articles
-
Mutations in the endodomain of Sindbis virus glycoprotein E2 define sequences critical for virus assembly.J Virol. 2006 May;80(9):4458-68. doi: 10.1128/JVI.80.9.4458-4468.2006. J Virol. 2006. PMID: 16611906 Free PMC article.
-
A single deletion in the membrane-proximal region of the Sindbis virus glycoprotein E2 endodomain blocks virus assembly.J Virol. 2000 May;74(9):4220-8. doi: 10.1128/jvi.74.9.4220-4228.2000. J Virol. 2000. PMID: 10756035 Free PMC article.
-
Single-Site Glycoprotein Mutants Inhibit a Late Event in Sindbis Virus Assembly.J Virol. 2016 Aug 26;90(18):8372-80. doi: 10.1128/JVI.00948-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27412592 Free PMC article.
-
Research on basis of reverse genetics system of a Sindbis-like virus XJ-160.Virol J. 2011 Nov 14;8:519. doi: 10.1186/1743-422X-8-519. Virol J. 2011. PMID: 22082202 Free PMC article. Review.
-
Molecular pathogenesis of Sindbis virus encephalitis in experimental animals.Adv Virus Res. 1989;36:255-71. doi: 10.1016/s0065-3527(08)60587-4. Adv Virus Res. 1989. PMID: 2544083 Review.
Cited by
-
Sindbis virus conformational changes induced by a neutralizing anti-E1 monoclonal antibody.J Virol. 2008 Jun;82(12):5750-60. doi: 10.1128/JVI.02673-07. Epub 2008 Apr 16. J Virol. 2008. PMID: 18417595 Free PMC article.
-
Chikungunya virus host range E2 transmembrane deletion mutants induce protective immunity against challenge in C57BL/6J mice.J Virol. 2013 Jun;87(12):6748-57. doi: 10.1128/JVI.03357-12. Epub 2013 Apr 3. J Virol. 2013. PMID: 23552427 Free PMC article.
-
Mutations in the endodomain of Sindbis virus glycoprotein E2 define sequences critical for virus assembly.J Virol. 2006 May;80(9):4458-68. doi: 10.1128/JVI.80.9.4458-4468.2006. J Virol. 2006. PMID: 16611906 Free PMC article.
-
Molecular links between the E2 envelope glycoprotein and nucleocapsid core in Sindbis virus.J Mol Biol. 2011 Dec 2;414(3):442-59. doi: 10.1016/j.jmb.2011.09.045. Epub 2011 Oct 4. J Mol Biol. 2011. PMID: 22001018 Free PMC article.
-
Interactions of the cytoplasmic domain of Sindbis virus E2 with nucleocapsid cores promote alphavirus budding.J Virol. 2012 Mar;86(5):2585-99. doi: 10.1128/JVI.05860-11. Epub 2011 Dec 21. J Virol. 2012. PMID: 22190727 Free PMC article.
References
-
- Anthony, R. P., A. M. Paredes, and D. T. Brown. 1992. Disulfide bonds are essential for the stability of the Sindbis virus envelope. Virology 190:330-336. - PubMed
-
- Braun, P., and G. von Heijne. 1999. The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry 38:9778-9782. - PubMed
-
- Bretscher, M. S. 1993. Cholesterol and the Golgi apparatus. Science 261:1280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

