Disruption of human TRIM5alpha antiviral activity by nonhuman primate orthologues
- PMID: 15919943
- PMCID: PMC1143641
- DOI: 10.1128/JVI.79.12.7883-7888.2005
Disruption of human TRIM5alpha antiviral activity by nonhuman primate orthologues
Abstract
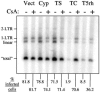
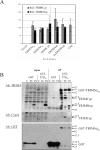
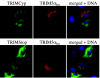
TRIM5 is a determinant of species-specific differences in susceptibility to infection by retroviruses bearing particular capsids. Human immunodeficiency virus type 1 (HIV-1) infection is blocked by the alpha isoform of macaque TRIM5 (TRIM5alpha(rh)) or by the product of the owl monkey TRIM5-cyclophilin A gene fusion (TRIMCyp). Human TRIM5alpha potently restricts specific strains of murine leukemia virus (N-MLV) but has only a modest effect on HIV-1. The amino termini of TRIM5 orthologues are highly conserved and possess a coiled-coil domain that promotes homomultimerization. Here we show that heterologous expression of TRIM5alpha(rh) or TRIMCyp in human cells interferes with the anti-N-MLV activity of endogenous human TRIM5alpha (TRIM5alpha(hu)). Deletion of the cyclophilin domain from TRIMCyp has no effect on heteromultimerization or colocalization with TRIM5alpha(hu) but prevents interference with anti-N-MLV activity. These data demonstrate that TRIM5 orthologues form heteromultimers and indicate that C-terminal extensions alter virus recognition by multimers of these proteins.
Figures


References
-
- Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11:286-291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

