The morphology and composition of influenza A virus particles are not affected by low levels of M1 and M2 proteins in infected cells
- PMID: 15919950
- PMCID: PMC1143655
- DOI: 10.1128/JVI.79.12.7926-7932.2005
The morphology and composition of influenza A virus particles are not affected by low levels of M1 and M2 proteins in infected cells
Abstract
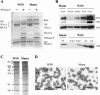
We generated a recombinant influenza A virus (Mmut) that produced low levels of matrix (M1) and M2 proteins in infected cells. Mmut virus propagated to significantly lower titers than did wild-type virus in cells infected at low multiplicity. By contrast, virion morphology and incorporation of viral proteins and vRNAs into virus particles were similar to those of wild-type virus. We propose that a threshold amount of M1 protein is needed for the assembly of viral components into an infectious particle and that budding is delayed in Mmut virus-infected cells until sufficient levels of M1 protein accumulate at the plasma membrane.
Figures
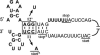

References
-
- Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. - PMC - PubMed
-
- Baudin, F., I. Petit, W. Weissenhorn, and R. W. Ruigrok. 2001. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 281:102-108. - PubMed
-
- Bourmakina, S. V., and A. Garcia-Sastre. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517-527. - PubMed
-
- Catchpole, A. P., L. J. Mingay, E. Fodor, and G. G. Brownlee. 2003. Alternative base pairs attenuate influenza A virus when introduced into the duplex region of the conserved viral RNA promoter of either the NS or the PA gene. J. Gen. Virol. 84:507-515. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

