Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD
- PMID: 15920478
- PMCID: PMC1150887
- DOI: 10.1038/sj.emboj.7600693
Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD
Abstract
Adhesive type 1 pili from uropathogenic Escherichia coli are filamentous protein complexes that are attached to the assembly platform FimD in the outer membrane. During pilus assembly, FimD binds complexes between the chaperone FimC and type 1 pilus subunits in the periplasm and mediates subunit translocation to the cell surface. Here we report nuclear magnetic resonance and X-ray protein structures of the N-terminal substrate recognition domain of FimD (FimD(N)) before and after binding of a chaperone-subunit complex. FimD(N) consists of a flexible N-terminal segment of 24 residues, a structured core with a novel fold, and a C-terminal hinge segment. In the ternary complex, residues 1-24 of FimD(N) specifically interact with both FimC and the subunit, acting as a sensor for loaded FimC molecules. Together with in vivo complementation studies, we show how this mechanism enables recognition and discrimination of different chaperone-subunit complexes by bacterial pilus assembly platforms.
Figures
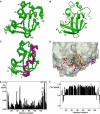
 (Pellecchia et al, 1999). (F) Heteronuclear [15N,1H]NOE measurements of FimDN(1–139) in the FimDN–FimC–FimHP ternary complex. Values between 0.5 and 1 indicate well-structured parts of the protein; values<0.5 manifest increased flexibility.
(Pellecchia et al, 1999). (F) Heteronuclear [15N,1H]NOE measurements of FimDN(1–139) in the FimDN–FimC–FimHP ternary complex. Values between 0.5 and 1 indicate well-structured parts of the protein; values<0.5 manifest increased flexibility.

Similar articles
-
Crystal structure of the ternary FimC-FimF(t)-FimD(N) complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD.FEBS Lett. 2008 Mar 5;582(5):651-5. doi: 10.1016/j.febslet.2008.01.030. Epub 2008 Jan 31. FEBS Lett. 2008. PMID: 18242189
-
Identification and characterization of the chaperone-subunit complex-binding domain from the type 1 pilus assembly platform FimD.J Mol Biol. 2003 Jul 11;330(3):513-25. doi: 10.1016/s0022-2836(03)00591-6. J Mol Biol. 2003. PMID: 12842468
-
Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD.Nature. 2018 Oct;562(7727):444-447. doi: 10.1038/s41586-018-0587-z. Epub 2018 Oct 3. Nature. 2018. PMID: 30283140 Free PMC article.
-
Chaperone-assisted assembly and molecular architecture of adhesive pili.Annu Rev Microbiol. 1991;45:383-415. doi: 10.1146/annurev.mi.45.100191.002123. Annu Rev Microbiol. 1991. PMID: 1683764 Review.
-
The molecular dissection of the chaperone-usher pathway.Biochim Biophys Acta. 2014 Aug;1843(8):1559-67. doi: 10.1016/j.bbamcr.2013.09.023. Epub 2013 Oct 16. Biochim Biophys Acta. 2014. PMID: 24140205 Review.
Cited by
-
Structure and function of enterotoxigenic Escherichia coli fimbriae from differing assembly pathways.Mol Microbiol. 2015 Jan;95(1):116-26. doi: 10.1111/mmi.12847. Epub 2014 Nov 27. Mol Microbiol. 2015. PMID: 25355550 Free PMC article.
-
Structure, Function, and Assembly of Adhesive Organelles by Uropathogenic Bacteria.Microbiol Spectr. 2015 Oct;3(5):10.1128/microbiolspec.UTI-0018-2013. doi: 10.1128/microbiolspec.UTI-0018-2013. Microbiol Spectr. 2015. PMID: 26542038 Free PMC article. Review.
-
Use of a combined cryo-EM and X-ray crystallography approach to reveal molecular details of bacterial pilus assembly by the chaperone/usher pathway.Curr Opin Microbiol. 2009 Jun;12(3):326-32. doi: 10.1016/j.mib.2009.03.002. Epub 2009 Apr 6. Curr Opin Microbiol. 2009. PMID: 19356973 Free PMC article. Review.
-
Pili Assembled by the Chaperone/Usher Pathway in Escherichia coli and Salmonella.EcoSal Plus. 2018 Mar;8(1):10.1128/ecosalplus.ESP-0007-2017. doi: 10.1128/ecosalplus.ESP-0007-2017. EcoSal Plus. 2018. PMID: 29536829 Free PMC article. Review.
-
Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria.EMBO J. 2008 Sep 3;27(17):2271-80. doi: 10.1038/emboj.2008.155. EMBO J. 2008. PMID: 18668121 Free PMC article. Review.
References
-
- Baorto DM, Gao Z, Malaviya R, Dustin ML, van der Merwe A, Lublin DM, Abraham SN (1997) Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature 389: 636–639 - PubMed
-
- Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K (1995) The program XEASY for computer-supported NMR spectral-analysis of biological macromolecules. J Biomol NMR 6: 1–10 - PubMed
-
- Barton GJ (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng 6: 37–40 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

