Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast
- PMID: 15920482
- PMCID: PMC1150880
- DOI: 10.1038/sj.emboj.7600683
Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast
Abstract
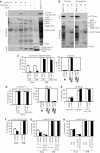
In eukaryotes, entry into mitosis is induced by cyclin B-bound Cdk1, which is held in check by the protein kinase, Wee1. In budding yeast, Swe1 (Wee1 ortholog) is targeted to the bud neck through Hsl1 (Nim1-related kinase) and its adaptor Hsl7, and is hyperphosphorylated prior to ubiquitin-mediated degradation. Here, we show that Hsl1 and Hsl7 are required for proper localization of Cdc5 (Polo-like kinase homolog) to the bud neck and Cdc5-dependent Swe1 phosphorylation. Mitotic cyclin (Clb2)-bound Cdc28 (Cdk1 homolog) directly phosphorylated Swe1 and this modification served as a priming step to promote subsequent Cdc5-dependent Swe1 hyperphosphorylation and degradation. Clb2-Cdc28 also facilitated Cdc5 localization to the bud neck through the enhanced interaction between the Clb2-Cdc28-phosphorylated Swe1 and the polo-box domain of Cdc5. We propose that the concerted action of Cdc28/Cdk1 and Cdc5/Polo on their common substrates is an evolutionarily conserved mechanism that is crucial for effectively triggering mitotic entry and other critical mitotic events.
Figures

References
-
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395–401 - PubMed
-
- Elia AE, Cantley LC, Yaffe MB (2003) Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299: 1228–1231 - PubMed
-
- Hwang WW, Silver PA (2001) The Saccharomyces cerevisiae cyclin Clb2p is targeted to multiple subcellular locations by cis- and trans-acting determinants. J Cell Sci 114: 589–597 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

